Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2
- PMID: 32547882
- PMCID: PMC7275676
- DOI: 10.7717/peerj.9278
Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2
Abstract
Background: Highly sensitive real-time reverse transcription polymerase chain reaction (RT-qPCR) methods have been developed for the detection of SARS-CoV-2. However, they are costly. Loop-mediated isothermal amplification (LAMP) assay has emerged as a novel alternative isothermal amplification method for the detection of nucleic acid.
Methods: A rapid, sensitive and specific real-time reverse transcription LAMP (RT-LAMP) assay was developed for SARS-CoV-2 detection.
Results: This assay detected one copy/reaction of SARS-CoV-2 RNA in 30 min. Both the clinical sensitivity and specificity of this assay were 100%. The RT-LAMP showed comparable performance with RT-qPCR. Combining simplicity and cost-effectiveness, this assay is therefore recommended for use in resource resource-limited settings.
Keywords: COVID-19; Diagnosis; RT-LAMP; Rapid detection.
© 2020 Lau et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
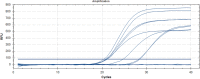
Similar articles
-
A Multiplex and Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Sensitive and Rapid Detection of Novel SARS-CoV-2.Front Cell Infect Microbiol. 2021 Jun 29;11:653616. doi: 10.3389/fcimb.2021.653616. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34268131 Free PMC article.
-
Development and Clinical Application of a Rapid and Sensitive Loop-Mediated Isothermal Amplification Test for SARS-CoV-2 Infection.mSphere. 2020 Aug 26;5(4):e00808-20. doi: 10.1128/mSphere.00808-20. mSphere. 2020. PMID: 32848011 Free PMC article.
-
A New Method to Detect Variants of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification Combined with a Bioluminescent Assay in Real Time (RT-LAMP-BART).Int J Mol Sci. 2023 Jun 27;24(13):10698. doi: 10.3390/ijms241310698. Int J Mol Sci. 2023. PMID: 37445876 Free PMC article.
-
Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic.Biology (Basel). 2020 Jul 22;9(8):182. doi: 10.3390/biology9080182. Biology (Basel). 2020. PMID: 32707972 Free PMC article. Review.
-
Reverse Transcriptase Loop Mediated Isothermal Amplification (RT-LAMP) for COVID-19 diagnosis: a systematic review and meta-analysis.Pathog Glob Health. 2021 Jul;115(5):281-291. doi: 10.1080/20477724.2021.1933335. Epub 2021 Jun 4. Pathog Glob Health. 2021. PMID: 34086539 Free PMC article.
Cited by
-
Rapid visual detection of SARS-CoV-2 by colorimetric loop-mediated isothermal amplification.Biotechniques. 2021 Apr;70(4):218-225. doi: 10.2144/btn-2020-0159. Epub 2021 Apr 6. Biotechniques. 2021. PMID: 33820475 Free PMC article.
-
A Simple, Affordable, Rapid, Stabilized, Colorimetric, Versatile RT-LAMP Assay to Detect SARS-CoV-2.Diagnostics (Basel). 2021 Mar 4;11(3):438. doi: 10.3390/diagnostics11030438. Diagnostics (Basel). 2021. PMID: 33806456 Free PMC article.
-
Loop-mediated isothermal amplification (LAMP): An effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2.Rev Med Virol. 2021 Nov;31(6):e2215. doi: 10.1002/rmv.2215. Epub 2021 Jan 21. Rev Med Virol. 2021. PMID: 33476080 Free PMC article. Review.
-
A Rapid RT-LAMP Assay for SARS-CoV-2 with Colorimetric Detection Assisted by a Mobile Application.Diagnostics (Basel). 2022 Mar 29;12(4):848. doi: 10.3390/diagnostics12040848. Diagnostics (Basel). 2022. PMID: 35453896 Free PMC article.
-
Review of COVID-19 testing and diagnostic methods.Talanta. 2022 Jul 1;244:123409. doi: 10.1016/j.talanta.2022.123409. Epub 2022 Mar 31. Talanta. 2022. PMID: 35390680 Free PMC article. Review.
References
-
- Cardoso TC, Ferrari HF, Bregano LC, Silva-Frade C, Rosa ACG, Andrade AL. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Molecular and Cellular Probes. 2010;24(6):415–417. doi: 10.1016/j.mcp.2010.08.003. - DOI - PMC - PubMed
-
- Centers for Disease Control Real-time RT-PCR panel for detection 2019-novel Coronavirus. 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-for-det.... [22 March 2020]. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-for-det...
-
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, Van Der Veer B, Van Den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
-
- Hu S-F, Li M, Zhong L-L, Lu S-M, Liu Z-X, Pu J-Y, Wen J-S, Huang X. Development of reverse-transcription loop-mediated isothermal amplification assay for rapid detection and differentiation of dengue virus serotypes 1–4. BMC Microbiology. 2015;15(1):265. doi: 10.1186/s12866-015-0595-1. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

