A Fully Automated Deep Learning Network for Brain Tumor Segmentation
- PMID: 32548295
- PMCID: PMC7289260
- DOI: 10.18383/j.tom.2019.00026
A Fully Automated Deep Learning Network for Brain Tumor Segmentation
Abstract
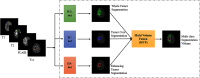
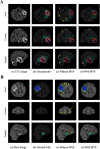
We developed a fully automated method for brain tumor segmentation using deep learning; 285 brain tumor cases with multiparametric magnetic resonance images from the BraTS2018 data set were used. We designed 3 separate 3D-Dense-UNets to simplify the complex multiclass segmentation problem into individual binary-segmentation problems for each subcomponent. We implemented a 3-fold cross-validation to generalize the network's performance. The mean cross-validation Dice-scores for whole tumor (WT), tumor core (TC), and enhancing tumor (ET) segmentations were 0.92, 0.84, and 0.80, respectively. We then retrained the individual binary-segmentation networks using 265 of the 285 cases, with 20 cases held-out for testing. We also tested the network on 46 cases from the BraTS2017 validation data set, 66 cases from the BraTS2018 validation data set, and 52 cases from an independent clinical data set. The average Dice-scores for WT, TC, and ET were 0.90, 0.84, and 0.80, respectively, on the 20 held-out testing cases. The average Dice-scores for WT, TC, and ET on the BraTS2017 validation data set, the BraTS2018 validation data set, and the clinical data set were as follows: 0.90, 0.80, and 0.78; 0.90, 0.82, and 0.80; and 0.85, 0.80, and 0.77, respectively. A fully automated deep learning method was developed to segment brain tumors into their subcomponents, which achieved high prediction accuracy on the BraTS data set and on the independent clinical data set. This method is promising for implementation into a clinical workflow.
Keywords: BraTS; Brain tumor segmentation; CNN (convolutional neural networks); Dense UNet; MRI; deep learning; machine learning.
© 2020 The Authors. Published by Grapho Publications, LLC.
Conflict of interest statement
Conflict of Interest: None reported
Figures

References
-
- Shreyas V, Pankajakshan V. A deep learning architecture for brain tumor segmentation in MRI images. In: 2017 IEEE 19th International Workshop on Multimedia Signal Processing (MMSP), Luton; 2017:1–6.
-
- Kamnitsas K, Bai W, Ferrante E, McDonagh S, Sinclair M, Pawlowski N, Rajchl M, Lee M, Kainz B, Rueckert D, Glocker B. Ensembles of multiple models and architectures for robust brain tumour segmentation. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, eds, Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Lecture notes in computer science, Vol. 10670 Springer: Cham; 2018:450–462.
-
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34:1993–2024. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

