Draft genome of the famous ornamental plant Paeonia suffruticosa
- PMID: 32551041
- PMCID: PMC7297784
- DOI: 10.1002/ece3.5965
Draft genome of the famous ornamental plant Paeonia suffruticosa
Abstract
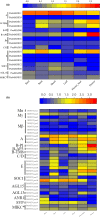
Tree peony (Paeonia Sect. Moutan) is a famous ornamental plant, with huge historical, cultural, and economic significance worldwide. In this study, we reported the ~13.79 Gb draft genome of a wide-grown Paeonia suffruticosa cultivar "Luo shen xiao chun," representing the largest sequenced genome in dicots to date. Phylogenetic analyses based on genome sequences demonstrated that P. suffruticosa was placed as sister to Vitales, and they together formed a clade that was sister to Rosids, weakly supporting a relationship of ((Saxifragales and Vitales) and Rosids). The identification and expression analysis of MADS-box genes based on the genome assembly and de novo transcriptome assembly of P. suffruticosa revealed that the function of C class genes was restricted in flower development, which might be responsible for the stamen petalody in tree peony cultivars. Overall, the first sequenced genome in the family Paeoniaceae provides an important resource for the origin, domestication, and evolutionary study as well as cultivar breeding in tree peony.
Keywords: Comparative genomics; MADS‐box; Paeonia suffruticosa; Tree peony; draft genome.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Figures


References
LinkOut - more resources
Full Text Sources

