DeeperBind: Enhancing Prediction of Sequence Specificities of DNA Binding Proteins
- PMID: 32551184
- PMCID: PMC7302108
- DOI: 10.1109/bibm.2016.7822515
DeeperBind: Enhancing Prediction of Sequence Specificities of DNA Binding Proteins
Abstract
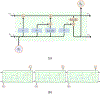
Transcription factors (TFs) are macromolecules that bind to cis-regulatory specific sub-regions of DNA promoters and initiate transcription. Finding the exact location of these binding sites (aka motifs) is important in a variety of domains such as drug design and development. To address this need, several in vivo and in vitro techniques have been developed so far that try to characterize and predict the binding specificity of a protein to different DNA loci. The major problem with these techniques is that they are not accurate enough in prediction of the binding affinity and characterization of the corresponding motifs. As a result, downstream analysis is required to uncover the locations where proteins of interest bind. Here, we propose DeeperBind, a long short term recurrent convolutional network for prediction of protein binding specificities with respect to DNA probes. DeeperBind can model the positional dynamics of probe sequences and hence reckons with the contributions made by individual sub-regions in DNA sequences, in an effective way. Moreover, it can be trained and tested on datasets containing varying-length sequences. We apply our pipeline to the datasets derived from protein binding microarrays (PBMs), an in-vitro high-throughput technology for quantification of protein-DNA binding preferences, and present promising results. To the best of our knowledge, this is the most accurate pipeline that can predict binding specificities of DNA sequences from the data produced by high-throughput technologies through utilization of the power of deep learning for feature generation and positional dynamics modeling.
Figures





References
-
- Alipanahi Babak, Delong Andrew, Matthew T Weirauch, and Brendan J Frey. Predicting the sequence specificities of dna-and rna-binding proteins by deep learning. Nature biotechnology, 2015. - PubMed
-
- Chen Xiaoyu, Hughes Timothy R., and Morris Quaid. RankMotif++: a motif-search algorithm that accounts for relative ranks of K-mers in binding transcription factors. Bioinformatics, 23(13):i72–i79, 2007. - PubMed
-
- Donahue Jeffrey, Lisa Anne Hendricks Sergio Guadarrama, Rohrbach Marcus, Venugopalan Subhashini, Saenko Kate, and Darrell Trevor. Long-term recurrent convolutional networks for visual recognition and description. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 2625–2634, 2015. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
