Modeling of nitrogen fixation and polymer production in the heterotrophic diazotroph Azotobacter vinelandii DJ
- PMID: 32551229
- PMCID: PMC7292883
- DOI: 10.1016/j.mec.2020.e00132
Modeling of nitrogen fixation and polymer production in the heterotrophic diazotroph Azotobacter vinelandii DJ
Abstract
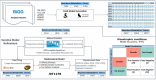
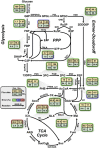
Nitrogen fixation is an important metabolic process carried out by microorganisms, which converts molecular nitrogen into inorganic nitrogenous compounds such as ammonia (NH3). These nitrogenous compounds are crucial for biogeochemical cycles and for the synthesis of essential biomolecules, i.e. nucleic acids, amino acids and proteins. Azotobacter vinelandii is a bacterial non-photosynthetic model organism to study aerobic nitrogen fixation (diazotrophy) and hydrogen production. Moreover, the diazotroph can produce biopolymers like alginate and polyhydroxybutyrate (PHB) that have important industrial applications. However, many metabolic processes such as partitioning of carbon and nitrogen metabolism in A. vinelandii remain unknown to date. Genome-scale metabolic models (M-models) represent reliable tools to unravel and optimize metabolic functions at genome-scale. M-models are mathematical representations that contain information about genes, reactions, metabolites and their associations. M-models can simulate optimal reaction fluxes under a wide variety of conditions using experimentally determined constraints. Here we report on the development of a M-model of the wild type bacterium A. vinelandii DJ (iDT1278) which consists of 2,003 metabolites, 2,469 reactions, and 1,278 genes. We validated the model using high-throughput phenotypic and physiological data, testing 180 carbon sources and 95 nitrogen sources. iDT1278 was able to achieve an accuracy of 89% and 91% for growth with carbon sources and nitrogen source, respectively. This comprehensive M-model will help to comprehend metabolic processes associated with nitrogen fixation, ammonium assimilation, and production of organic nitrogen in an environmentally important microorganism.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Castillo T., Heinzle E., Peifer S., Schneider K., Peña M.C.F. Oxygen supply strongly influences metabolic fluxes, the production of poly(3-hydroxybutyrate) and alginate, and the degree of acetylation of alginate in Azotobacter vinelandii. Process Biochem. 2013;48:995–1003. doi: 10.1016/j.procbio.2013.04.014. - DOI
-
- Chang R.L., Ghamsari L., Manichaikul A., Hom E.F.Y., Balaji S., Fu W., Shen Y., Hao T., Palsson B.Ø., Salehi-Ashtiani K., Papin J.A. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 2011;7:518. doi: 10.1038/msb.2011.52. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

