Validation of Free Energy Methods in AMBER
- PMID: 32551593
- PMCID: PMC7686255
- DOI: 10.1021/acs.jcim.0c00285
Validation of Free Energy Methods in AMBER
Abstract
Herein we provide high-precision validation tests of the latest GPU-accelerated free energy code in AMBER. We demonstrate that consistent free energy results are obtained in both the gas phase and in solution. We first show, in the context of thermodynamic integration (TI), that the results are invariant with respect to "split" (e.g., stepwise decharge-vdW-recharge) versus "unified" protocols. This brought to light a subtle inconsistency in previous versions of AMBER that was traced to the improper treatment of 1-4 vdW and electrostatic interactions involving atoms across the softcore boundary. We illustrate that under the assumption that the ensembles produced by different legs of the alchemical transformation between molecules A and B in the gas phase and aqueous phase are very small, the inconsistency in the relative hydration free energy ΔΔGhydr[A → B] = ΔGaq[A → B] - ΔGgas[A → B] is minimal. However, for general cases where the ensembles are shown to be substantially different, as expected in ligand-protein binding applications, these errors can be large. Finally, we demonstrate that results for relative hydration free energy simulations are independent of TI or multistate Bennett's acceptance ratio (MBAR) analysis, invariant to the specific choice of the softcore region, and in agreement with results derived from absolute hydration free energy values.
Figures

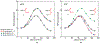
References
-
- Chipot C Frontiers in free-energy calculations of biological systems. Wiley Interdisciplinary Reviews: Computational Molecular Science 2014, 4, 71–89.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

