Machine Learning Classifier Models Can Identify Acute Respiratory Distress Syndrome Phenotypes Using Readily Available Clinical Data
- PMID: 32551817
- PMCID: PMC7528785
- DOI: 10.1164/rccm.202002-0347OC
Machine Learning Classifier Models Can Identify Acute Respiratory Distress Syndrome Phenotypes Using Readily Available Clinical Data
Abstract
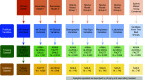
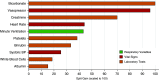
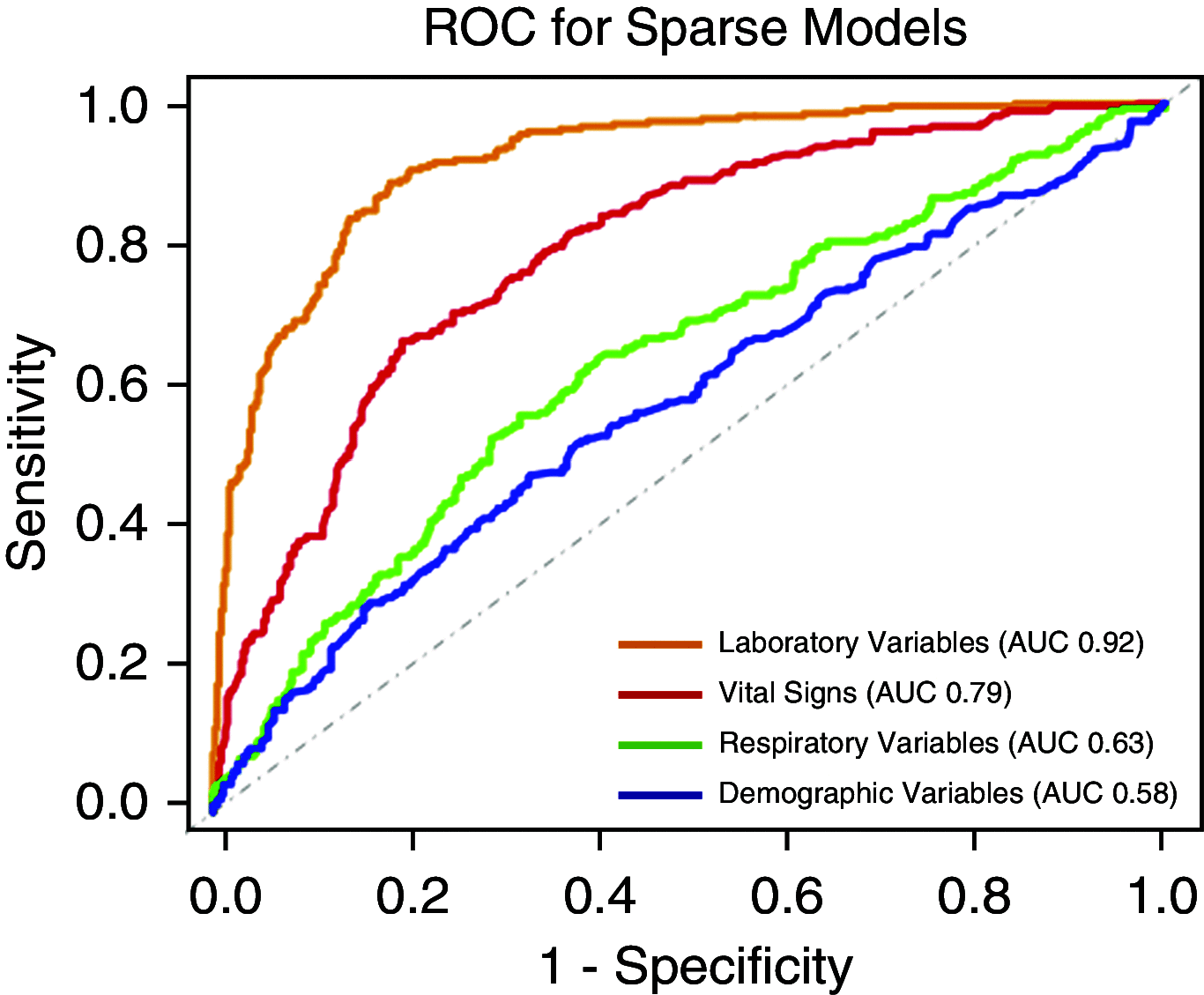
Rationale: Two distinct phenotypes of acute respiratory distress syndrome (ARDS) with differential clinical outcomes and responses to randomly assigned treatment have consistently been identified in randomized controlled trial cohorts using latent class analysis. Plasma biomarkers, key components in phenotype identification, currently lack point-of-care assays and represent a barrier to the clinical implementation of phenotypes.Objectives: The objective of this study was to develop models to classify ARDS phenotypes using readily available clinical data only.Methods: Three randomized controlled trial cohorts served as the training data set (ARMA [High vs. Low Vt], ALVEOLI [Assessment of Low Vt and Elevated End-Expiratory Pressure to Obviate Lung Injury], and FACTT [Fluids and Catheter Treatment Trial]; n = 2,022), and a fourth served as the validation data set (SAILS [Statins for Acutely Injured Lungs from Sepsis]; n = 745). A gradient-boosted machine algorithm was used to develop classifier models using 24 variables (demographics, vital signs, laboratory, and respiratory variables) at enrollment. In two secondary analyses, the ALVEOLI and FACTT cohorts each, individually, served as the validation data set, and the remaining combined cohorts formed the training data set for each analysis. Model performance was evaluated against the latent class analysis-derived phenotype.Measurements and Main Results: For the primary analysis, the model accurately classified the phenotypes in the validation cohort (area under the receiver operating characteristic curve [AUC], 0.95; 95% confidence interval [CI], 0.94-0.96). Using a probability cutoff of 0.5 to assign class, inflammatory biomarkers (IL-6, IL-8, and sTNFR-1; P < 0.0001) and 90-day mortality (38% vs. 24%; P = 0.0002) were significantly higher in the hyperinflammatory phenotype as classified by the model. Model accuracy was similar when ALVEOLI (AUC, 0.94; 95% CI, 0.92-0.96) and FACTT (AUC, 0.94; 95% CI, 0.92-0.95) were used as the validation cohorts. Significant treatment interactions were observed with the clinical classifier model-assigned phenotypes in both ALVEOLI (P = 0.0113) and FACTT (P = 0.0072) cohorts.Conclusions: ARDS phenotypes can be accurately identified using machine learning models based on readily available clinical data and may enable rapid phenotype identification at the bedside.
Keywords: ARDS phenotypes; classifier models; machine learning.
Figures
Comment in
-
Machine Learning Classifier Models: The Future for Acute Respiratory Distress Syndrome Phenotyping?Am J Respir Crit Care Med. 2020 Oct 1;202(7):919-920. doi: 10.1164/rccm.202006-2388ED. Am J Respir Crit Care Med. 2020. PMID: 32687397 Free PMC article. No abstract available.
References
-
- Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. LUNG SAFE Investigators; ESICM Trials Group. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315:788–800. - PubMed
-
- Kumar G, Kumar N, Taneja A, Kaleekal T, Tarima S, McGinley E, et al. Milwaukee Initiative in Critical Care Outcomes Research (MICCOR) Group of Investigators. Nationwide trends of severe sepsis in the 21st century (2000-2007) Chest. 2011;140:1223–1231. - PubMed
-
- Matthay MA, McAuley DF, Ware LB. Clinical trials in acute respiratory distress syndrome: challenges and opportunities. Lancet Respir Med. 2017;5:524–534. - PubMed
-
- Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20:195–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

