Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics
- PMID: 32552793
- PMCID: PMC7298854
- DOI: 10.1186/s13059-020-02053-9
Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics
Abstract
Background: At least 50% of patients with suspected Mendelian disorders remain undiagnosed after whole-exome sequencing (WES), and the extent to which non-coding variants that are not captured by WES contribute to this fraction is unclear. Whole transcriptome sequencing is a promising supplement to WES, although empirical data on the contribution of RNA analysis to the diagnosis of Mendelian diseases on a large scale are scarce.
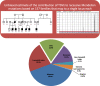
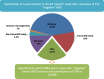
Results: Here, we describe our experience with transcript-deleterious variants (TDVs) based on a cohort of 5647 families with suspected Mendelian diseases. We first interrogate all families for which the respective Mendelian phenotype could be mapped to a single locus to obtain an unbiased estimate of the contribution of TDVs at 18.9%. We examine the entire cohort and find that TDVs account for 15% of all "solved" cases. We compare the results of RT-PCR to in silico prediction. Definitive results from RT-PCR are obtained from blood-derived RNA for the overwhelming majority of variants (84.1%), and only a small minority (2.6%) fail analysis on all available RNA sources (blood-, skin fibroblast-, and urine renal epithelial cells-derived), which has important implications for the clinical application of RNA-seq. We also show that RNA analysis can establish the diagnosis in 13.5% of 155 patients who had received "negative" clinical WES reports. Finally, our data suggest a role for TDVs in modulating penetrance even in otherwise highly penetrant Mendelian disorders.
Conclusions: Our results provide much needed empirical data for the impending implementation of diagnostic RNA-seq in conjunction with genome sequencing.
Keywords: Mapping; Mendelian; Negative WES; RNA-based diagnostics; Transcriptomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

