Novel venom-based peptides (P13 and its derivative-M6) to maintain self-renewal of human embryonic stem cells by activating FGF and TGFβ signaling pathways
- PMID: 32552810
- PMCID: PMC7302175
- DOI: 10.1186/s13287-020-01766-9
Novel venom-based peptides (P13 and its derivative-M6) to maintain self-renewal of human embryonic stem cells by activating FGF and TGFβ signaling pathways
Abstract
Background: In our previous study, a venom-based peptide named Gonearrestide (also named P13) was identified and demonstrated with an effective inhibition in the proliferation of colon cancer cells. In this study, we explored if P13 and its potent mutant M6 could promote the proliferation of human embryonic stem cells and even maintain their self-renewal.
Methods: The structure-function relationship analysis on P13 and its potent mutant M6 were explored from the molecular mechanism of corresponding receptor activation by a series of inhibitor assay plus molecular and dynamics simulation studies.
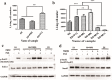
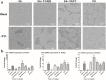
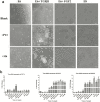
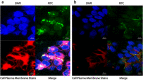
Results: An interesting phenomenon is that P13 (and its potent mutant M6), an 18AA short peptide, can activate both FGF and TGFβ signaling pathways. We demonstrated that the underlying molecular mechanisms of P13 and M6 could cooperate with proteoglycans to complete the "dimerization" of FGFR and TGFβ receptors.
Conclusions: Taken together, this study is the first research finding on a venom-based peptide that works on the FGF and TGF-β signaling pathways to maintain the self-renewal of hESCs.
Keywords: Embryonic stem cell; Peptide modification; Pluripotency; Self-renewal; Venom peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- King GF. Venoms as a platform for human drugs: translating toxins into therapeutics. Expert Opin Biol Ther. 2011;11(11):1469–1484. - PubMed
-
- King GF, Escoubas P, Nicholson GM. Peptide toxins that selectively target insect Na-V and Ca-V channels. Channels. 2008;2(2):100–116. - PubMed
-
- de la Vega RC R, Possani LD. Overview of scorpion toxins specific for Na+ channels and related peptides: biodiversity, structure-function relationships and evolution. Toxicon. 2005;46(8):831–844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

