Identification of novel cerebrospinal fluid biomarker candidates for dementia with Lewy bodies: a proteomic approach
- PMID: 32552841
- PMCID: PMC7301448
- DOI: 10.1186/s13024-020-00388-2
Identification of novel cerebrospinal fluid biomarker candidates for dementia with Lewy bodies: a proteomic approach
Abstract
Background: Diagnosis of dementia with Lewy bodies (DLB) is challenging, largely due to a lack of diagnostic tools. Cerebrospinal fluid (CSF) biomarkers have been proven useful in Alzheimer's disease (AD) diagnosis. Here, we aimed to identify novel CSF biomarkers for DLB using a high-throughput proteomic approach.
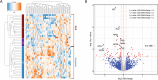
Methods: We applied liquid chromatography/tandem mass spectrometry with label-free quantification to identify biomarker candidates to individual CSF samples from a well-characterized cohort comprising patients with DLB (n = 20) and controls (n = 20). Validation was performed using (1) the identical proteomic workflow in an independent cohort (n = 30), (2) proteomic data from patients with related neurodegenerative diseases (n = 149) and (3) orthogonal techniques in an extended cohort consisting of DLB patients and controls (n = 76). Additionally, we utilized random forest analysis to identify the subset of candidate markers that best distinguished DLB from all other groups.
Results: In total, we identified 1995 proteins. In the discovery cohort, 69 proteins were differentially expressed in DLB compared to controls (p < 0.05). Independent cohort replication confirmed VGF, SCG2, NPTX2, NPTXR, PDYN and PCSK1N as candidate biomarkers for DLB. The downregulation of the candidate biomarkers was somewhat more pronounced in DLB in comparison with related neurodegenerative diseases. Using random forest analysis, we identified a panel of VGF, SCG2 and PDYN to best differentiate between DLB and other clinical groups (accuracy: 0.82 (95%CI: 0.75-0.89)). Moreover, we confirmed the decrease of VGF and NPTX2 in DLB by ELISA and SRM methods. Low CSF levels of all biomarker candidates, except PCSK1N, were associated with more pronounced cognitive decline (0.37 < r < 0.56, all p < 0.01).
Conclusion: We identified and validated six novel CSF biomarkers for DLB. These biomarkers, particularly when used as a panel, show promise to improve diagnostic accuracy and strengthen the importance of synaptic dysfunction in the pathophysiology of DLB.
Keywords: Biomarkers; Cerebrospinal fluid; Dementia with Lewy bodies; Lewy body dementia; Proteomics.
Conflict of interest statement
Nothing to report.
Figures




References
-
- Vekrellis K, Xilouri M, Emmanouilidou E, Rideout HJ, Stefanis L. Pathological roles of alpha-synuclein in neurological disorders. Lancet Neurol. 2011;10(11):1015–1025. - PubMed
-
- Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6(3):131–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

