Dynamic linkage of COVID-19 test results between Public Health England's Second Generation Surveillance System and UK Biobank
- PMID: 32553051
- PMCID: PMC7478634
- DOI: 10.1099/mgen.0.000397
Dynamic linkage of COVID-19 test results between Public Health England's Second Generation Surveillance System and UK Biobank
Abstract
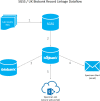
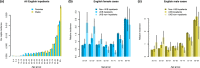
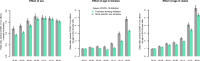
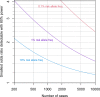
UK Biobank (UKB) is an international health resource enabling research into the genetic and lifestyle determinants of common diseases of middle and older age. It comprises 500 000 participants. Public Health England's Second Generation Surveillance System is a centralized microbiology database covering English clinical diagnostics laboratories that provides national surveillance of legally notifiable infections, bacterial isolations and antimicrobial resistance. We previously developed secure, pseudonymized, individual-level linkage of these systems. In this study, we implemented rapid dynamic linkage, which allows us to provide a regular feed of new COVID-19 (SARS-CoV-2) test results to UKB to facilitate rapid and urgent research into the epidemiological and human genetic risk factors for severe infection in the cohort. Here, we have characterized the first 1352 cases of COVID-19 in UKB participants, of whom 895 met our working definition of severe COVID-19 as inpatients hospitalized on or after 16 March 2020. We found that the incidence of severe COVID-19 among UKB cases was 27.4 % lower than the general population in England, although this difference varied significantly by age and sex. The total number of UKB cases could be estimated as 0.6 % of the publicly announced number of cases in England. We considered how increasing case numbers will affect the power of genome-wide association studies. This new dynamic linkage system has further potential to facilitate the investigation of other infections and the prospective collection of microbiological cultures to create a microbiological biobank (bugbank) for studying the interaction of environment, human and microbial genetics on infection in the UKB cohort.
Keywords: COVID-19; Public Health England; SARS-CoV-2; UK Biobank; bugbank; database linkage.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Chinese Preventive Medicine Association An update on the epidemiological characteristics of novel coronavirus pneumonia (COVID-19) Chin J Epidemiol. 2020;41:139–144. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

