tRNAArg-Derived Fragments Can Serve as Arginine Donors for Protein Arginylation
- PMID: 32553119
- PMCID: PMC7409373
- DOI: 10.1016/j.chembiol.2020.05.013
tRNAArg-Derived Fragments Can Serve as Arginine Donors for Protein Arginylation
Abstract
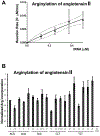
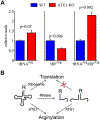
Arginyltransferase ATE1 mediates posttranslational arginylation and plays key roles in multiple physiological processes. ATE1 utilizes arginyl (Arg)-tRNAArg as the donor of Arg, putting this reaction into a direct competition with the protein synthesis machinery. Here, we address the question of ATE1- Arg-tRNAArg specificity as a potential mechanism enabling this competition in vivo. Using in vitro arginylation assays and Ate1 knockout models, we find that, in addition to full-length tRNA, ATE1 is also able to utilize short tRNAArg fragments that bear structural resemblance to tRNA-derived fragments (tRF), a recently discovered class of small regulatory non-coding RNAs with global emerging biological role. Ate1 knockout cells show a decrease in tRFArg generation and a significant increase in the ratio of tRNAArg:tRFArg compared with wild type, suggesting a functional link between tRFArg and arginylation. We propose that generation of physiologically important tRFs can serve as a switch between translation and protein arginylation.
Keywords: arginylation; tRF; tRNA.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


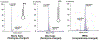

References
-
- Barciszewska MZ, Erdmann VA, and Barciszewski J (1996). Ribosomal 5S RNA: tertiary structure and interactions with proteins. Biological reviews of the Cambridge Philosophical Society 71, 1–25. - PubMed
-
- Beckert B, Turk M, Czech A, Berninghausen O, Beckmann R, Ignatova Z, Plitzko JM, and Wilson DN (2018). Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nature microbiology 3, 1115–1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

