Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis
- PMID: 32553350
- PMCID: PMC7245202
- DOI: 10.1016/j.bios.2020.112316
Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis
Abstract
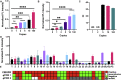
Recent research suggests that SARS-CoV-2-infected individuals can be highly infectious while asymptomatic or pre-symptomatic, and that an infected person may infect 5.6 other individuals on average. This situation highlights the need for rapid, sensitive SARS-CoV-2 diagnostic assays capable of high-throughput operation that can preferably utilize existing equipment to facilitate broad, large-scale screening efforts. We have developed a CRISPR-based assay that can meet all these criteria. This assay utilizes a custom CRISPR Cas12a/gRNA complex and a fluorescent probe to detect target amplicons produced by standard RT-PCR or isothermal recombinase polymerase amplification (RPA), to allow sensitive detection at sites not equipped with real-time PCR systems required for qPCR diagnostics. We found this approach allowed sensitive and robust detection of SARS-CoV-2 positive samples, with a sample-to-answer time of ~50 min, and a limit of detection of 2 copies per sample. CRISPR assay diagnostic results obtained nasal swab samples of individuals with suspected COVID-19 cases were comparable to paired results from a CDC-approved quantitative RT-PCR (RT-qPCR) assay performed in a state testing lab, and superior to those produced by same assay in a clinical lab, where the RT-qPCR assay exhibited multiple invalid or inconclusive results. Our assay also demonstrated greater analytical sensitivity and more robust diagnostic performance than other recently reported CRISPR-based assays. Based on these findings, we believe that a CRISPR-based fluorescent application has potential to improve current COVID-19 screening efforts.
Keywords: COVID-19; CRISPR; Fluorescent detection; Highly sensitive; Molecular diagnosis; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Bachman J. Reverse-transcription PCR (RT-PCR) Methods Enzymol. 2013;530:67–74. - PubMed
-
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020:1–5. doi: 10.1038/s41587-020-0513-4. - DOI - PMC - PubMed
-
- Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a-Powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31(51):1905311. - PubMed
-
- Bruch R., Urban G.A., Dincer C. CRISPR/Cas powered multiplexed biosensing. Trends Biotechnol. 2019;37(8):791–792. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

