The selective PPAR-delta agonist seladelpar reduces ethanol-induced liver disease by restoring gut barrier function and bile acid homeostasis in mice
- PMID: 32553670
- PMCID: PMC7719076
- DOI: 10.1016/j.trsl.2020.06.006
The selective PPAR-delta agonist seladelpar reduces ethanol-induced liver disease by restoring gut barrier function and bile acid homeostasis in mice
Abstract
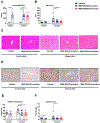
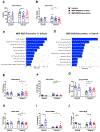
Alcohol-associated liver disease is accompanied by dysregulation of bile acid metabolism and gut barrier dysfunction. Peroxisome proliferator-activated receptor-delta (PPARδ) agonists are key metabolic regulators and have anti-inflammatory properties. Here, we evaluated the effect of the selective PPAR-delta agonist seladelpar (MBX-8025) on gut barrier function and bile acid metabolism in a mouse model of ethanol-induced liver disease. Wild type C57BL/6 mice were fed LieberDeCarli diet containing 0%-36% ethanol (caloric) for 8 weeks followed by a single binge of ethanol (5 g/kg). Pair fed mice received an isocaloric liquid diet as control. MBX-8025 (10 mg/kg/d) or vehicle were added to the liquid diet during the entire feeding period (prevention), or during the last 4 weeks of Lieber DeCarli diet feeding (intervention). In both prevention and intervention trials, MBX-8025 protected mice from ethanol-induced liver disease, characterized by lower serum alanine aminotransferase (ALT) levels, hepatic triglycerides, and inflammation. Chronic ethanol intake disrupted bile acid metabolism by increasing the total bile acid pool and serum bile acids. MBX-8025 reduced serum total and secondary bile acids, and the total bile acid pool as compared with vehicle treatment in both prevention and intervention trials. MBX-8025 restored ethanol-induced gut dysbiosis and gut barrier dysfunction. Data from this study demonstrates that seladelpar prevents and treats ethanol-induced liver damage in mice by direct PPARδ agonism in both the liver and the intestine.
Keywords: Enterohepatic circulation; Gut barrier; Microbiome.
Copyright © 2020 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Effects of elafibranor on liver fibrosis and gut barrier function in a mouse model of alcohol-associated liver disease.World J Gastroenterol. 2024 Jul 28;30(28):3428-3446. doi: 10.3748/wjg.v30.i28.3428. World J Gastroenterol. 2024. PMID: 39091710 Free PMC article.
-
New strain of Pediococcus pentosaceus alleviates ethanol-induced liver injury by modulating the gut microbiota and short-chain fatty acid metabolism.World J Gastroenterol. 2020 Oct 28;26(40):6224-6240. doi: 10.3748/wjg.v26.i40.6224. World J Gastroenterol. 2020. PMID: 33177795 Free PMC article.
-
Fibroblast Growth Factor 19 Alters Bile Acids to Induce Dysbiosis in Mice With Alcohol-Induced Liver Disease.Cell Mol Gastroenterol Hepatol. 2024;18(1):71-87. doi: 10.1016/j.jcmgh.2024.02.015. Epub 2024 Feb 28. Cell Mol Gastroenterol Hepatol. 2024. PMID: 38417701 Free PMC article.
-
n-3 Polyunsaturated fatty acids for the management of alcoholic liver disease: A critical review.Crit Rev Food Sci Nutr. 2019;59(sup1):S116-S129. doi: 10.1080/10408398.2018.1544542. Epub 2018 Dec 22. Crit Rev Food Sci Nutr. 2019. PMID: 30580553 Review.
-
Bile Acid Metabolism in Liver Pathobiology.Gene Expr. 2018 May 18;18(2):71-87. doi: 10.3727/105221618X15156018385515. Epub 2018 Jan 11. Gene Expr. 2018. PMID: 29325602 Free PMC article. Review.
Cited by
-
Effects of elafibranor on liver fibrosis and gut barrier function in a mouse model of alcohol-associated liver disease.World J Gastroenterol. 2024 Jul 28;30(28):3428-3446. doi: 10.3748/wjg.v30.i28.3428. World J Gastroenterol. 2024. PMID: 39091710 Free PMC article.
-
Transcriptomic Profiling Identifies Novel Hepatic and Intestinal Genes Following Chronic Plus Binge Ethanol Feeding in Mice.Dig Dis Sci. 2020 Dec;65(12):3592-3604. doi: 10.1007/s10620-020-06461-6. Epub 2020 Jul 15. Dig Dis Sci. 2020. PMID: 32671585 Free PMC article.
-
Peroxisome proliferator-activated receptor agonists: A new hope towards the management of alcoholic liver disease.World J Gastroenterol. 2024 Sep 21;30(35):3965-3971. doi: 10.3748/wjg.v30.i35.3965. World J Gastroenterol. 2024. PMID: 39351059 Free PMC article.
-
Gram-positive probiotics improves acetaminophen-induced hepatotoxicity by inhibiting leucine and Hippo-YAP pathway.Cell Biosci. 2025 Mar 7;15(1):32. doi: 10.1186/s13578-025-01370-5. Cell Biosci. 2025. PMID: 40055814 Free PMC article.
-
Key Signaling in Alcohol-Associated Liver Disease: The Role of Bile Acids.Cells. 2022 Apr 18;11(8):1374. doi: 10.3390/cells11081374. Cells. 2022. PMID: 35456053 Free PMC article. Review.
References
-
- Crabb DW, Im GY, Szabo G, Mellinger JL, Lucey MR. Diagnosis and treatment of alcohol-associated liver diseases: 2019 practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2020;71:306–33. - PubMed
-
- Dang K, Hirode G, Singal AK, Sundaram V, Wong RJ. Alcoholic liver disease epidemiology in the United States: a retrospective analysis of 3 US databases. Am J Gastroenterol 2020;115:96–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

