Cytochrome P4501B1 in bone marrow is co-expressed with key markers of mesenchymal stem cells. BMS2 cell line models PAH disruption of bone marrow niche development functions
- PMID: 32553695
- PMCID: PMC7293885
- DOI: 10.1016/j.taap.2020.115111
Cytochrome P4501B1 in bone marrow is co-expressed with key markers of mesenchymal stem cells. BMS2 cell line models PAH disruption of bone marrow niche development functions
Abstract
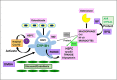
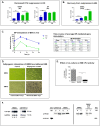
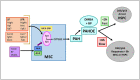
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous pollutants that are metabolized to carcinogenic dihydrodiol epoxides (PAHDE) by cytochrome P450 1B1 (CYP1B1). This metabolism occurs in bone marrow (BM) mesenchymal stem cells (MSC), which sustain hematopoietic stem and progenitor cells (HSPC). In BM, CYP1B1-mediated metabolism of 7, 12-dimethylbenz[a]anthracene (DMBA) suppresses HSPC colony formation within 6 h, whereas benzo(a)pyrene (BP) generates protective cytokines. MSC, enriched from adherent BM cells, yielded the bone marrow stromal, BMS2, cell line. These cells express elevated basal CYP1B1 that scarcely responds to Ah receptor (AhR) inducers. BMS2 cells exhibit extensive transcriptome overlap with leptin receptor positive mesenchymal stem cells (Lepr+ MSC) that control the hematopoietic niche. The overlap includes CYP1B1 and the expression of HSPC regulatory factors (Ebf3, Cxcl12, Kitl, Csf1 and Gas6). MSC are large, adherent fibroblasts that sequester small HSPC and macrophage in the BM niche (Graphic abstract). High basal CYP1B1 expression in BMS2 cells derives from interactions between the Ah-receptor enhancer and proximal promoter SP1 complexes, boosted by autocrine signaling. PAH effects on BMS2 cells model Lepr+MSC niche activity. CYP1B1 metabolizes DMBA to PAHDE, producing p53-mediated mRNA increases, long after the in vivo HSPC suppression. Faster, direct p53 effects, favored by stem cells, remain possible PAHDE targets. However, HSPC regulatory factors remained unresponsive. BP is less toxic in BMS2 cells, but, in BM, CYP1A1 metabolism stimulates macrophage cytokines (Il1b > Tnfa> Ifng) within 6 h. Although absent from BMS2 and Lepr+MSC, their receptors are highly expressed. The impact of this cytokine signaling in MSC remains to be determined.
Keywords: BMS2 cells; Bone marrow vascular niche; CYP1B1; Hematopoietic stem and progenitor cells; Lepr+MSC; Mesenchymal stem cells.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors acknowledge no conflicts of interest with these studies.
Figures






Similar articles
-
AhR and CYP1B1 Control Oxygen Effects on Bone Marrow Progenitor Cells: The Enrichment of Multiple Olfactory Receptors as Potential Microbiome Sensors.Int J Mol Sci. 2023 Nov 28;24(23):16884. doi: 10.3390/ijms242316884. Int J Mol Sci. 2023. PMID: 38069208 Free PMC article.
-
Bone marrow stromal cells constitutively express high levels of cytochrome P4501B1 that metabolize 7,12-dimethylbenz[a]anthracene.Mol Pharmacol. 1998 Dec;54(6):1000-6. doi: 10.1124/mol.54.6.1000. Mol Pharmacol. 1998. PMID: 9855628
-
Regulation of preB cell apoptosis by aryl hydrocarbon receptor/transcription factor-expressing stromal/adherent cells.Proc Soc Exp Biol Med. 1999 Jul;221(3):242-52. doi: 10.3181/00379727-221-44410. Proc Soc Exp Biol Med. 1999. PMID: 10404042
-
From Marrow to Bone and Fat: Exploring the Multifaceted Roles of Leptin Receptor Positive Bone Marrow Mesenchymal Stromal Cells.Cells. 2024 May 24;13(11):910. doi: 10.3390/cells13110910. Cells. 2024. PMID: 38891042 Free PMC article. Review.
-
Hematopoietic Niche - Exploring Biomimetic Cues to Improve the Functionality of Hematopoietic Stem/Progenitor Cells.Biotechnol J. 2018 Feb;13(2). doi: 10.1002/biot.201700088. Epub 2017 Dec 28. Biotechnol J. 2018. PMID: 29178199 Review.
Cited by
-
Defined Diets Link Iron and α-Linolenic Acid to Cyp1b1 Regulation of Neonatal Liver Development Through Srebp Forms and LncRNA H19.Int J Mol Sci. 2025 Feb 25;26(5):2011. doi: 10.3390/ijms26052011. Int J Mol Sci. 2025. PMID: 40076634 Free PMC article.
-
AhR and CYP1B1 Control Oxygen Effects on Bone Marrow Progenitor Cells: The Enrichment of Multiple Olfactory Receptors as Potential Microbiome Sensors.Int J Mol Sci. 2023 Nov 28;24(23):16884. doi: 10.3390/ijms242316884. Int J Mol Sci. 2023. PMID: 38069208 Free PMC article.
-
Fruit Extract, Rich in Polyphenols and Flavonoids, Modifies the Expression of DNMT and HDAC Genes Involved in Epigenetic Processes.Nutrients. 2023 Apr 13;15(8):1867. doi: 10.3390/nu15081867. Nutrients. 2023. PMID: 37111085 Free PMC article.
-
Association between urinary polycyclic aromatic hydrocarbon metabolites and premature menopause: a nationally representative cross-sectional study in the United States.Environ Health Prev Med. 2025;30:32. doi: 10.1265/ehpm.25-00031. Environ Health Prev Med. 2025. PMID: 40335317 Free PMC article.
References
-
- Alexander D.L., Eltom S.E., Jefcoate C.R. Ah receptor regulation of CYP1B1 expression in primary mouse embryo-derived cells. Cancer Res. 1997;57(20):4498–4506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

