LC-FACSeq is a method for detecting rare clones in leukemia
- PMID: 32554930
- PMCID: PMC7406301
- DOI: 10.1172/jci.insight.134973
LC-FACSeq is a method for detecting rare clones in leukemia
Abstract
Detecting, characterizing, and monitoring rare populations of cells can increase testing sensitivity, give insight into disease mechanism, and inform clinical decision making. One area that can benefit from increased resolution is management of cancers in clinical remission but with measurable residual disease (MRD) by multicolor FACS. Detecting and monitoring genomic clonal resistance to treatment in the setting of MRD is technically difficult and resource intensive due to the limited amounts of disease cells. Here, we describe limited-cell FACS sequencing (LC-FACSeq), a reproducible, highly sensitive method of characterizing clonal evolution in rare cells relevant to different types of acute and chronic leukemias. We demonstrate the utility of LC-FACSeq for broad multigene gene panels and its application for monitoring sequential acquisition of mutations conferring therapy resistance and clonal evolution in long-term ibrutinib treatment of patients with chronic lymphocytic leukemia. This technique is generalizable for monitoring of other blood and marrow infiltrating cancers.
Keywords: Clonal selection; Diagnostics; Genetics; Leukemias.
Conflict of interest statement
Figures
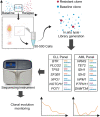
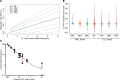


References
-
- Hallek M, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. - DOI - PMC - PubMed
-
- van der Velden VH, Hochhaus A, Cazzaniga G, Szczepanski T, Gabert J, van Dongen JJ. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003;17(6):1013–1034. doi: 10.1038/sj.leu.2402922. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

