Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome
- PMID: 32554932
- PMCID: PMC7406263
- DOI: 10.1172/jci.insight.135678
Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome
Abstract
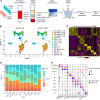
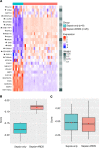
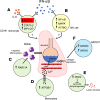
Acute respiratory distress syndrome (ARDS) results from overwhelming pulmonary inflammation. Prior bulk RNA sequencing provided limited insights into ARDS pathogenesis. We used single cell RNA sequencing to probe ARDS at a higher resolution. PBMCs of patients with pneumonia and sepsis with early ARDS were compared with those of sepsis patients who did not develop ARDS. Monocyte clusters from ARDS patients revealed multiple distinguishing characteristics in comparison with monocytes from patients without ARDS, including downregulation of SOCS3 expression, accompanied by a proinflammatory signature with upregulation of multiple type I IFN-induced genes, especially in CD16+ cells. To generate an ARDS risk score, we identified upregulation of 29 genes in the monocytes of these patients, and 17 showed a similar profile in cells of patients in independent cohorts. Monocytes had increased expression of RAB11A, known to inhibit neutrophil efferocytosis; ATP2B1, a calcium pump that exports Ca2+ implicated in endothelial barrier disruption; and SPARC, associated with processing of procollagen to collagen. These data show that monocytes of ARDS patients upregulate expression of genes not just restricted to those associated with inflammation. Together, our findings identify molecules that are likely involved in ARDS pathogenesis that may inform biomarker and therapeutic development.
Keywords: Immunology; Innate immunity; Pulmonology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

