Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution
- PMID: 32555182
- PMCID: PMC7300015
- DOI: 10.1038/s41467-020-16876-4
Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution
Abstract
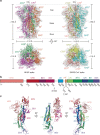
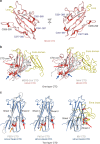
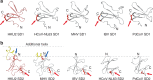
Porcine coronavirus SADS-CoV has been identified from suckling piglets with severe diarrhea in southern China in 2017. The SADS-CoV genome shares ~95% identity to that of bat α-coronavirus HKU2, suggesting that SADS-CoV may have emerged from a natural reservoir in bats. Here we report the cryo-EM structures of HKU2 and SADS-CoV spike (S) glycoprotein trimers at 2.38 Å and 2.83 Å resolution, respectively. We systematically compare the domains of HKU2 spike with those of α-, β-, γ-, and δ-coronavirus spikes, showing that the S1 subunit N- and C-terminal domains of HKU2/SADS-CoV are ancestral domains in the evolution of coronavirus spike proteins. The connecting region after the fusion peptide in the S2 subunit of HKU2/SADS-CoV adopts a unique conformation. These results structurally demonstrate a close evolutionary relationship between HKU2/SADS-CoV and β-coronavirus spikes and provide insights into the evolution and cross-species transmission of coronaviruses.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.J Virol. 2020 Oct 27;94(22):e01301-20. doi: 10.1128/JVI.01301-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32817223 Free PMC article.
-
Broad Cross-Species Infection of Cultured Cells by Bat HKU2-Related Swine Acute Diarrhea Syndrome Coronavirus and Identification of Its Replication in Murine Dendritic Cells In Vivo Highlight Its Potential for Diverse Interspecies Transmission.J Virol. 2019 Nov 26;93(24):e01448-19. doi: 10.1128/JVI.01448-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554686 Free PMC article.
-
The origin and evolution of emerged swine acute diarrhea syndrome coronavirus with zoonotic potential.J Med Virol. 2023 Mar;95(3):e28672. doi: 10.1002/jmv.28672. J Med Virol. 2023. PMID: 36916779
-
Interplay of swine acute diarrhoea syndrome coronavirus and the host intrinsic and innate immunity.Vet Res. 2025 Jan 9;56(1):5. doi: 10.1186/s13567-024-01436-1. Vet Res. 2025. PMID: 39789633 Free PMC article. Review.
-
Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery.Virus Res. 2020 Aug;285:198024. doi: 10.1016/j.virusres.2020.198024. Epub 2020 May 16. Virus Res. 2020. PMID: 32482591 Free PMC article. Review.
Cited by
-
Characterization and Spike Gene Analysis of a Candidate Attenuated Live Bovine Coronavirus Vaccine.Animals (Basel). 2024 Jan 25;14(3):389. doi: 10.3390/ani14030389. Animals (Basel). 2024. PMID: 38338032 Free PMC article.
-
Establishment of replication-competent vesicular stomatitis virus recapitulating SADS-CoV entry.J Virol. 2024 May 14;98(5):e0195723. doi: 10.1128/jvi.01957-23. Epub 2024 Apr 1. J Virol. 2024. PMID: 38557247 Free PMC article.
-
Update on the Phylodynamics of SADS-CoV.Life (Basel). 2021 Aug 11;11(8):820. doi: 10.3390/life11080820. Life (Basel). 2021. PMID: 34440564 Free PMC article.
-
PF-00835231 broadly inhibits swine Alpha-coronavirus, including emerging SADS-CoV.J Virol. 2024 Nov 19;98(11):e0130324. doi: 10.1128/jvi.01303-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480086 Free PMC article.
-
Functional binding dynamics relevant to the evolution of zoonotic spillovers in endemic and emergent Betacoronavirus strains.bioRxiv [Preprint]. 2021 Apr 20:2020.09.11.293258. doi: 10.1101/2020.09.11.293258. bioRxiv. 2021. Update in: J Biomol Struct Dyn. 2022;40(21):10978-10996. doi: 10.1080/07391102.2021.1953604. PMID: 33501438 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

