Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis
- PMID: 32555190
- PMCID: PMC7303144
- DOI: 10.1038/s41419-020-2581-2
Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis
Abstract
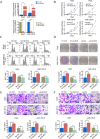
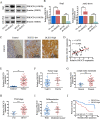
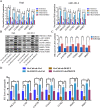
Long noncoding RNAs (lncRNAs) have been identified as potential prognostic tools and therapeutic biomarkers for a variety of human cancers. However, the functional roles and underlying mechanisms of key lncRNAs affecting laryngeal squamous cell carcinomas (LSCCs) are largely unknown. Here, we adopted a novel subpathway strategy based on the lncRNA-mRNA profiles from the Cancer Genome Atlas (TCGA) database and identified the lncRNA deleted in lymphocytic leukemia 2 (DLEU2) as an oncogene in the pathogenesis of LSCCs. We found that DLEU2 was significantly upregulated and predicted poor clinical outcomes in LSCC patients. In addition, ectopic overexpression of DLEU2 promoted the proliferation and migration of LSCC cells both in vivo and in vitro. Mechanistically, DLEU2 served as a competing endogenous RNA to regulate PIK3CD expression by sponging miR-30c-5p and subsequently activated the Akt signaling pathway. As a target gene of DLEU2, PIK3CD was also upregulated and could predict a poor prognosis in LSCC patients. In conclusion, we found that the novel LSCC-related gene DLEU2 enhances the malignant properties of LSCCs via the miR-30c-5p/PIK3CD/Akt axis. DLEU2 and its targeted miR-30c-5p/PIK3CD/Akt axis may represent valuable prognostic biomarkers and therapeutic targets for LSCCs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. - PubMed
-
- Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J. Clin. 2017;67:31–50. - PubMed
-
- Bates JE, et al. Curative-dose chemoradiotherapy versus total laryngectomy for stage T3−T4 squamous cell carcinoma of the larynx: an “apples-to-apples” analysis of the National Cancer Database. Am. J. Clin. Oncol. 2019;42:527–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

