Genomic deregulation of PRMT5 supports growth and stress tolerance in chronic lymphocytic leukemia
- PMID: 32555249
- PMCID: PMC7299935
- DOI: 10.1038/s41598-020-66224-1
Genomic deregulation of PRMT5 supports growth and stress tolerance in chronic lymphocytic leukemia
Abstract
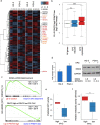
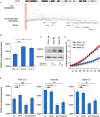
Patients suffering from chronic lymphocytic leukemia (CLL) display highly diverse clinical courses ranging from indolent cases to aggressive disease, with genetic and epigenetic features resembling this diversity. Here, we developed a comprehensive approach combining a variety of molecular and clinical data to pinpoint translocation events disrupting long-range chromatin interactions and causing cancer-relevant transcriptional deregulation. Thereby, we discovered a B cell specific cis-regulatory element restricting the expression of genes in the associated locus, including PRMT5 and DAD1, two factors with oncogenic potential. Experimental PRMT5 inhibition identified transcriptional programs similar to those in patients with differences in PRMT5 abundance, especially MYC-driven and stress response pathways. In turn, such inhibition impairs factors involved in DNA repair, sensitizing cells for apoptosis. Moreover, we show that artificial deletion of the regulatory element from its endogenous context resulted in upregulation of corresponding genes, including PRMT5. Furthermore, such disruption renders PRMT5 transcription vulnerable to additional stimuli and subsequently alters the expression of downstream PRMT5 targets. These studies provide a mechanism of PRMT5 deregulation in CLL and the molecular dependencies identified might have therapeutic implementations.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth.Oncogene. 2017 Mar 2;36(9):1223-1231. doi: 10.1038/onc.2016.287. Epub 2016 Aug 22. Oncogene. 2017. PMID: 27546619 Free PMC article.
-
Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells.Mol Cell Biol. 2008 Oct;28(20):6262-77. doi: 10.1128/MCB.00923-08. Epub 2008 Aug 11. Mol Cell Biol. 2008. PMID: 18694959 Free PMC article.
-
Targeting PRMT5/Akt signalling axis prevents human lung cancer cell growth.J Cell Mol Med. 2019 Feb;23(2):1333-1342. doi: 10.1111/jcmm.14036. Epub 2018 Nov 20. J Cell Mol Med. 2019. PMID: 30461193 Free PMC article.
-
Interaction between miRNAs and signaling cascades of Wnt pathway in chronic lymphocytic leukemia.J Cell Biochem. 2020 Nov;121(11):4654-4666. doi: 10.1002/jcb.29683. Epub 2020 Feb 26. J Cell Biochem. 2020. PMID: 32100920 Review.
-
Role of protein arginine methyltransferase 5 in human cancers.Biomed Pharmacother. 2019 Jun;114:108790. doi: 10.1016/j.biopha.2019.108790. Epub 2019 Mar 20. Biomed Pharmacother. 2019. PMID: 30903920 Review.
Cited by
-
Extracellular vesicles from type-2 macrophages increase the survival of chronic lymphocytic leukemia cells ex vivo.Cancer Gene Ther. 2024 Aug;31(8):1164-1176. doi: 10.1038/s41417-024-00802-7. Epub 2024 Jun 25. Cancer Gene Ther. 2024. PMID: 38918490 Free PMC article.
-
Protein Arginine Methyltransferases as Therapeutic Targets in Hematological Malignancies.Cancers (Basel). 2022 Nov 5;14(21):5443. doi: 10.3390/cancers14215443. Cancers (Basel). 2022. PMID: 36358861 Free PMC article. Review.
-
Emerging role of BAD and DAD1 as potential targets and biomarkers in cancer.Oncol Lett. 2021 Dec;22(6):811. doi: 10.3892/ol.2021.13072. Epub 2021 Sep 28. Oncol Lett. 2021. PMID: 34671425 Free PMC article. Review.
-
Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter's transformation.Nat Commun. 2023 Jan 6;14(1):97. doi: 10.1038/s41467-022-35778-1. Nat Commun. 2023. PMID: 36609611 Free PMC article.
-
ZNF224 Protein: Multifaceted Functions Based on Its Molecular Partners.Molecules. 2021 Oct 18;26(20):6296. doi: 10.3390/molecules26206296. Molecules. 2021. PMID: 34684876 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

