RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses
- PMID: 32555404
- PMCID: PMC7303170
- DOI: 10.1038/s41598-020-66735-x
RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses
Abstract
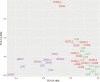
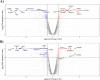
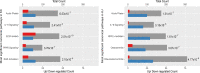
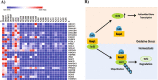
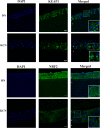
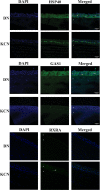
Keratoconus is a highly prevalent (1 in 2000), genetically complex and multifactorial, degenerative disease of the cornea whose pathogenesis and underlying transcriptomic changes are poorly understood. To identify disease-specific changes and gene expression networks, we performed next generation RNA sequencing from individual corneas of two distinct patient populations - one from the Middle East, as keratoconus is particularly severe in this group, and the second from an African American population in the United States. We conducted a case: control RNA sequencing study of 7 African American, 12 Middle Eastern subjects, and 7 controls. A Principal Component Analysis of all expressed genes was used to ascertain differences between samples. Differentially expressed genes were identified using Cuffdiff and DESeq2 analyses, and identification of over-represented signaling pathways by Ingenuity Pathway Analysis. Although separated by geography and ancestry, key commonalities in the two patient transcriptomes speak of disease - intrinsic gene expression networks. We identified an overwhelming decrease in the expression of anti-oxidant genes regulated by NRF2 and those of the acute phase and tissue injury response pathways, in both patient groups. Concordantly, NRF2 immunofluorescence staining was decreased in patient corneas, while KEAP1, which helps to degrade NRF2, was increased. Diminished NRF2 signaling raises the possibility of NRF2 activators as future treatment strategies in keratoconus. The African American patient group showed increases in extracellular matrix transcripts that may be due to underlying profibrogenic changes in this group. Transcripts increased across all patient samples include Thrombospondin 2 (THBS2), encoding a matricellular protein, and cellular proteins, GAS1, CASR and OTOP2, and are promising biomarker candidates. Our approach of analyzing transcriptomic data from different populations and patient groups will help to develop signatures and biomarkers for keratoconus subtypes. Further, RNA sequence data on individual patients obtained from multiple studies may lead to a core keratoconus signature of deregulated genes and a better understanding of its pathogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Krachmer, J. H., Feder, R. S. & Belin, M. W. Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol28, 293-322, 0039-6257(84)90094-8 [pii] (1984). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

