Acute inhibition of centriolar satellite function and positioning reveals their functions at the primary cilium
- PMID: 32555591
- PMCID: PMC7326281
- DOI: 10.1371/journal.pbio.3000679
Acute inhibition of centriolar satellite function and positioning reveals their functions at the primary cilium
Abstract
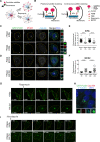
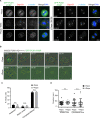
Centriolar satellites are dynamic, membraneless granules composed of over 200 proteins. They store, modify, and traffic centrosome and primary cilium proteins, and help to regulate both the biogenesis and some functions of centrosomes and cilium. In most cell types, satellites cluster around the perinuclear centrosome, but their integrity and cellular distribution are dynamically remodeled in response to different stimuli, such as cell cycle cues. Dissecting the specific and temporal functions and mechanisms of satellites and how these are influenced by their cellular positioning and dynamics has been challenging using genetic approaches, particularly in ciliated and proliferating cells. To address this, we developed a chemical-based trafficking assay to rapidly and efficiently redistribute satellites to either the cell periphery or center, and fuse them into stable clusters in a temporally controlled way. Induced satellite clustering at either the periphery or center resulted in antagonistic changes in the pericentrosomal levels of a subset of proteins, revealing a direct and selective role for their positioning in protein targeting and sequestration. Systematic analysis of the interactome of peripheral satellite clusters revealed enrichment of proteins implicated in cilium biogenesis and mitosis. Importantly, induction of peripheral satellite targeting in ciliated cells revealed a function for satellites not just for efficient cilium assembly but also in the maintenance of steady-state cilia and in cilia disassembly by regulating the structural integrity of the ciliary axoneme. Finally, perturbing satellite distribution and dynamics inhibited their mitotic dissolution, and mitotic progression was perturbed only in cells with centrosomal satellite clustering. Collectively, our results for the first time showed a direct link between satellite functions and their pericentrosomal clustering, suggested new mechanisms underlying satellite functions during cilium assembly, and provided a new tool for probing temporal satellite functions in different contexts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

