Identifying and predicting Parkinson's disease subtypes through trajectory clustering via bipartite networks
- PMID: 32555729
- PMCID: PMC7299311
- DOI: 10.1371/journal.pone.0233296
Identifying and predicting Parkinson's disease subtypes through trajectory clustering via bipartite networks
Abstract
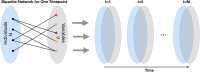
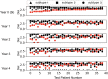
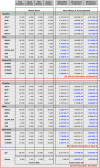
Chronic medical conditions show substantial heterogeneity in their clinical features and progression. We develop the novel data-driven, network-based Trajectory Profile Clustering (TPC) algorithm for 1) identification of disease subtypes and 2) early prediction of subtype/disease progression patterns. TPC is an easily generalizable method that identifies subtypes by clustering patients with similar disease trajectory profiles, based not only on Parkinson's Disease (PD) variable severity, but also on their complex patterns of evolution. TPC is derived from bipartite networks that connect patients to disease variables. Applying our TPC algorithm to a PD clinical dataset, we identify 3 distinct subtypes/patient clusters, each with a characteristic progression profile. We show that TPC predicts the patient's disease subtype 4 years in advance with 72% accuracy for a longitudinal test cohort. Furthermore, we demonstrate that other types of data such as genetic data can be integrated seamlessly in the TPC algorithm. In summary, using PD as an example, we present an effective method for subtype identification in multidimensional longitudinal datasets, and early prediction of subtypes in individual patients.
Conflict of interest statement
Data was obtained from the Parkinson’s Progression Markers Initiative (PPMI). PPMI, a public-private partnership, is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Allergan, Avid Radiopharmaceuticals, Biogen, Biolegend, Bristol-Myers Squibb, Celgene, Denali, GE Healthcare, Genentech, gsk, Lilly, Pfizer, Merck, MSD, Lundbeck, Piramal, Prevail Therapeutics, Roche, Sanofi Genzyme, Servier, Takeda, Teva, Ucb, Verily, Voyager Therapeutics and Golub Capital. There are no patents, products in development or marketed products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



References
-
- Lees AJ, Hardy J, Revesz T. Parkinson’s Disease. Lancet. 2009;374 (9691).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

