Genomewide Association Study of Severe Covid-19 with Respiratory Failure
- PMID: 32558485
- PMCID: PMC7315890
- DOI: 10.1056/NEJMoa2020283
Genomewide Association Study of Severe Covid-19 with Respiratory Failure
Abstract
Background: There is considerable variation in disease behavior among patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (Covid-19). Genomewide association analysis may allow for the identification of potential genetic factors involved in the development of Covid-19.
Methods: We conducted a genomewide association study involving 1980 patients with Covid-19 and severe disease (defined as respiratory failure) at seven hospitals in the Italian and Spanish epicenters of the SARS-CoV-2 pandemic in Europe. After quality control and the exclusion of population outliers, 835 patients and 1255 control participants from Italy and 775 patients and 950 control participants from Spain were included in the final analysis. In total, we analyzed 8,582,968 single-nucleotide polymorphisms and conducted a meta-analysis of the two case-control panels.
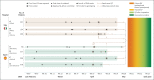
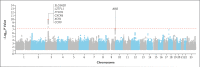
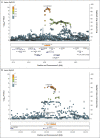
Results: We detected cross-replicating associations with rs11385942 at locus 3p21.31 and with rs657152 at locus 9q34.2, which were significant at the genomewide level (P<5×10-8) in the meta-analysis of the two case-control panels (odds ratio, 1.77; 95% confidence interval [CI], 1.48 to 2.11; P = 1.15×10-10; and odds ratio, 1.32; 95% CI, 1.20 to 1.47; P = 4.95×10-8, respectively). At locus 3p21.31, the association signal spanned the genes SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6 and XCR1. The association signal at locus 9q34.2 coincided with the ABO blood group locus; in this cohort, a blood-group-specific analysis showed a higher risk in blood group A than in other blood groups (odds ratio, 1.45; 95% CI, 1.20 to 1.75; P = 1.48×10-4) and a protective effect in blood group O as compared with other blood groups (odds ratio, 0.65; 95% CI, 0.53 to 0.79; P = 1.06×10-5).
Conclusions: We identified a 3p21.31 gene cluster as a genetic susceptibility locus in patients with Covid-19 with respiratory failure and confirmed a potential involvement of the ABO blood-group system. (Funded by Stein Erik Hagen and others.).
Copyright © 2020 Massachusetts Medical Society.
Figures
Comment in
-
Genetic Risk of Severe Covid-19.N Engl J Med. 2020 Oct 15;383(16):1590-1591. doi: 10.1056/NEJMe2025501. N Engl J Med. 2020. PMID: 33053291 Free PMC article. No abstract available.
-
ABO blood groups are not associated with risk of acquiring the SARS-CoV-2 infection in young adults.Haematologica. 2020 Dec 1;105(12):2841-2843. doi: 10.3324/haematol.2020.265066. Haematologica. 2020. PMID: 33256383 Free PMC article. No abstract available.
-
Mining a GWAS of Severe Covid-19.N Engl J Med. 2020 Dec 24;383(26):2588-2589. doi: 10.1056/NEJMc2025747. Epub 2020 Nov 24. N Engl J Med. 2020. PMID: 33289971 No abstract available.
-
Mining a GWAS of Severe Covid-19.N Engl J Med. 2020 Dec 24;383(26):2589. doi: 10.1056/NEJMc2025747. Epub 2020 Nov 24. N Engl J Med. 2020. PMID: 33289972 No abstract available.
-
DNA genotyping of the ABO gene showed a significant association of the A-group (A1/A2 variants) with severe COVID-19.Eur J Intern Med. 2021 Jun;88:129-132. doi: 10.1016/j.ejim.2021.02.016. Epub 2021 Feb 25. Eur J Intern Med. 2021. PMID: 33750629 Free PMC article. No abstract available.
-
Coagulation factors and the incidence of COVID-19 severity: Mendelian randomization analyses and supporting evidence.Signal Transduct Target Ther. 2021 Jun 7;6(1):222. doi: 10.1038/s41392-021-00640-1. Signal Transduct Target Ther. 2021. PMID: 34099622 Free PMC article. No abstract available.
References
-
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020. February 24 (Epub ahead of print). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
- Andalusian Health Repository - access to free full text
- Archivio Istituzionale della Ricerca Unimi - Access Free Full Text
- Atypon
- Diposit Digital de la Universitat de Barcelona - Access Free Full Text
- Europe PubMed Central
- Ovid Technologies, Inc.
- PubMed Central
- University of Turin Instituional Repository AperTO - Free full text
Other Literature Sources
Medical
Miscellaneous
