Isolation, Sequence, Infectivity, and Replication Kinetics of Severe Acute Respiratory Syndrome Coronavirus 2
- PMID: 32558639
- PMCID: PMC7454076
- DOI: 10.3201/eid2609.201495
Isolation, Sequence, Infectivity, and Replication Kinetics of Severe Acute Respiratory Syndrome Coronavirus 2
Abstract
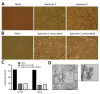
Since its emergence in Wuhan, China, in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected ≈6 million persons worldwide. As SARS-CoV-2 spreads across the planet, we explored the range of human cells that can be infected by this virus. We isolated SARS-CoV-2 from 2 infected patients in Toronto, Canada; determined the genomic sequences; and identified single-nucleotide changes in representative populations of our virus stocks. We also tested a wide range of human immune cells for productive infection with SARS-CoV-2. We confirm that human primary peripheral blood mononuclear cells are not permissive for SARS-CoV-2. As SARS-CoV-2 continues to spread globally, it is essential to monitor single-nucleotide polymorphisms in the virus and to continue to isolate circulating viruses to determine viral genotype and phenotype by using in vitro and in vivo infection models.
Keywords: 2019 novel coronavirus disease; COVID-19; SARS-CoV-2; coronavirus disease; immune cells; isolation; phylogenetics; replication; respiratory infections; severe acute respiratory syndrome coronavirus 2; viruses; zoonoses.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

