A Mechanistic, Enantioselective, Physiologically Based Pharmacokinetic Model of Verapamil and Norverapamil, Built and Evaluated for Drug-Drug Interaction Studies
- PMID: 32560124
- PMCID: PMC7355632
- DOI: 10.3390/pharmaceutics12060556
A Mechanistic, Enantioselective, Physiologically Based Pharmacokinetic Model of Verapamil and Norverapamil, Built and Evaluated for Drug-Drug Interaction Studies
Abstract
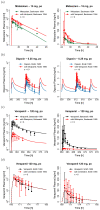
The calcium channel blocker and antiarrhythmic agent verapamil is recommended by the FDA for drug-drug interaction (DDI) studies as a moderate clinical CYP3A4 index inhibitor and as a clinical Pgp inhibitor. The purpose of the presented work was to develop a mechanistic whole-body physiologically based pharmacokinetic (PBPK) model to investigate and predict DDIs with verapamil. The model was established in PK-Sim®, using 45 clinical studies (dosing range 0.1-250 mg), including literature as well as unpublished Boehringer Ingelheim data. The verapamil R- and S-enantiomers and their main metabolites R- and S-norverapamil are represented in the model. The processes implemented to describe the pharmacokinetics of verapamil and norverapamil include enantioselective plasma protein binding, enantioselective metabolism by CYP3A4, non-stereospecific Pgp transport, and passive glomerular filtration. To describe the auto-inhibitory and DDI potential, mechanism-based inactivation of CYP3A4 and non-competitive inhibition of Pgp by the verapamil and norverapamil enantiomers were incorporated based on in vitro literature. The resulting DDI performance was demonstrated by prediction of DDIs with midazolam, digoxin, rifampicin, and cimetidine, with 21/22 predicted DDI AUC ratios or Ctrough ratios within 1.5-fold of the observed values. The thoroughly built and qualified model will be freely available in the Open Systems Pharmacology model repository to support model-informed drug discovery and development.
Keywords: P-glycoprotein (Pgp); cytochrome P450 3A4 (CYP3A4); drug–drug interactions (DDIs); mechanism-based inactivation (MBI); model-informed drug discovery and development (MID3); non-competitive inhibition; norverapamil; physiologically based pharmacokinetic (PBPK) modeling; verapamil.
Conflict of interest statement
Sabrina Wiebe, Éric Fernandez, Peter Stopfer, and Valerie Nock are employees of Boehringer Ingelheim Pharma GmbH & Co. KG. Thorsten Lehr has received research grants from Boehringer Ingelheim Pharma GmbH & Co. KG. and from the German Federal Ministry of Education and Research (grants 03XP0196 and 031L0161C). Nina Hanke, Denise Türk, and Dominik Selzer declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- ClinCalc LLC. ClinCalc DrugStats Database. [(accessed on 25 March 2020)]; Available online: https://clincalc.com/DrugStats/
-
- Belpomme D., Gauthier S., Pujade-Lauraine E., Facchini T., Goudier M.J., Krakowski I., Netter-Pinon G., Frenay M., Gousset C., Marié F.N., et al. Verapamil increases the survival of patients with anthracycline-resistant metastatic breast carcinoma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2000;11:1471–1476. doi: 10.1023/A:1026556119020. - DOI - PubMed
-
- Tolcher A.W., Cowan K.H., Solomon D., Ognibene F., Goldspiel B., Chang R., Noone M.H., Denicoff A.M., Barnes C.S., Gossard M.R., et al. Phase I crossover study of paclitaxel with r-verapamil in patients with metastatic breast cancer. J. Clin. Oncol. 1996;14:1173–1184. doi: 10.1200/JCO.1996.14.4.1173. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

