Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation
- PMID: 32561270
- PMCID: PMC7276322
- DOI: 10.1016/j.immuni.2020.06.001
Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation
Abstract
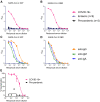
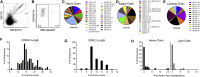
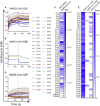
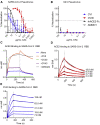
Antibody responses develop following SARS-CoV-2 infection, but little is known about their epitope specificities, clonality, binding affinities, epitopes, and neutralizing activity. We isolated B cells specific for the SARS-CoV-2 envelope glycoprotein spike (S) from a COVID-19-infected subject 21 days after the onset of clinical disease. 45 S-specific monoclonal antibodies were generated. They had undergone minimal somatic mutation with limited clonal expansion, and three bound the receptor-binding domain (RBD). Two antibodies neutralized SARS-CoV-2. The most potent antibody bound the RBD and prevented binding to the ACE2 receptor, while the other bound outside the RBD. Thus, most anti-S antibodies that were generated in this patient during the first weeks of COVID-19 infection were non-neutralizing and target epitopes outside the RBD. Antibodies that disrupt the SARS-CoV-2 S-ACE2 interaction can potently neutralize the virus without undergoing extensive maturation. Such antibodies have potential preventive and/or therapeutic potential and can serve as templates for vaccine design.
Keywords: ACE2; B cells; COVID-19; MERS; SARS; SARS-CoV-2; antibodies; neutralization; receptor-binding domain; spike protein.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing financial interests. A provisional patent application (U.S. Provisional Application No. 63/016268) has been filed on the SARS-CoV-2 specific monoclonal antibodies isolated herein. H.Y.C. receives personal fees from Merck (consultant), personal fees from Glaxo Smith Kline (consultant), grants from Sanofi-Pasteur, non-financial support from Cepheid, non-financial support from Ellume, and non-financial support from Genentech. The content of these consultancies and support are unrelated to the work performed in this manuscript.
Figures

Update of
-
Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual.bioRxiv [Preprint]. 2020 May 12:2020.05.12.091298. doi: 10.1101/2020.05.12.091298. bioRxiv. 2020. Update in: Immunity. 2020 Jul 14;53(1):98-105.e5. doi: 10.1016/j.immuni.2020.06.001. PMID: 32511342 Free PMC article. Updated. Preprint.
References
-
- Charif D., Lobry J. SeqinR 1.0-2: a contributed package to the R Project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U., Porto M., Roman H.E., Vendruscolo M., editors. Structural Approaches to Sequence Evolution. Springer Verlag; 2007. pp. 207–232.
-
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

