Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution
- PMID: 32561545
- PMCID: PMC7397856
- DOI: 10.1101/gad.330050.119
Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution
Abstract
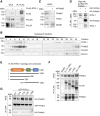
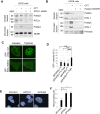
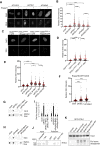
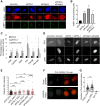
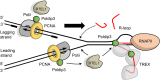
RTEL1 helicase is a component of DNA repair and telomere maintenance machineries. While RTEL1's role in DNA replication is emerging, how RTEL1 preserves genomic stability during replication remains elusive. Here we used a range of proteomic, biochemical, cell, and molecular biology and gene editing approaches to provide further insights into potential role(s) of RTEL1 in DNA replication and genome integrity maintenance. Our results from complementary human cell culture models established that RTEL1 and the Polδ subunit Poldip3 form a complex and are/function mutually dependent in chromatin binding after replication stress. Loss of RTEL1 and Poldip3 leads to marked R-loop accumulation that is confined to sites of active replication, enhances endogenous replication stress, and fuels ensuing genomic instability. The impact of depleting RTEL1 and Poldip3 is epistatic, consistent with our proposed concept of these two proteins operating in a shared pathway involved in DNA replication control under stress conditions. Overall, our data highlight a previously unsuspected role of RTEL1 and Poldip3 in R-loop suppression at genomic regions where transcription and replication intersect, with implications for human diseases including cancer.
Keywords: DNA damage response; DNA repair; DNA:RNA hybrid; Hoyeral-Hreiderson syndrome; POLδ; Poldip3; R-loop; RTEL1; dyskeratosis congenita; helicase; polymerase δ; telomere maintenance.
© 2020 Björkman et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Ballew BJ, Joseph V, De S, Sarek G, Vannier JB, Stracker T, Schrader KA, Small TN, O'Reilly R, Manschreck C, et al. 2013a. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet 9: e1003695 10.1371/journal.pgen.1003695 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
