The guanine nucleotide exchange factor VAV3 participates in ERBB4-mediated cancer cell migration
- PMID: 32561640
- PMCID: PMC7450113
- DOI: 10.1074/jbc.RA119.010925
The guanine nucleotide exchange factor VAV3 participates in ERBB4-mediated cancer cell migration
Abstract
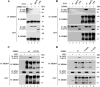
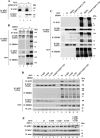
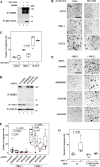
ERBB4 is a member of the epidermal growth factor receptor (EGFR)/ERBB subfamily of receptor tyrosine kinases that regulates cellular processes including proliferation, migration, and survival. ERBB4 signaling is involved in embryogenesis and homeostasis of healthy adult tissues, but also in human pathologies such as cancer, neurological disorders, and cardiovascular diseases. Here, an MS-based analysis revealed the Vav guanine nucleotide exchange factor 3 (VAV3), an activator of Rho family GTPases, as a critical ERBB4-interacting protein in breast cancer cells. We confirmed the ERBB4-VAV3 interaction by targeted MS and coimmunoprecipitation experiments and further defined it by demonstrating that kinase activity and Tyr-1022 and Tyr-1162 of ERBB4, as well as the intact phosphotyrosine-interacting SH2 domain of VAV3, are necessary for this interaction. We found that ERBB4 stimulates tyrosine phosphorylation of the VAV3 activation domain, known to be required for guanine nucleotide exchange factor (GEF) activity of VAV proteins. In addition to VAV3, the other members of the VAV family, VAV1 and VAV2, also coprecipitated with ERBB4. Analyses of the effects of overexpression of dominant-negative VAV3 constructs or shRNA-mediated down-regulation of VAV3 expression in breast cancer cells indicated that active VAV3 is involved in ERBB4-stimulated cell migration. These results define the VAV GEFs as effectors of ERBB4 activity in a signaling pathway relevant for cancer cell migration.
Keywords: ERBB4; HER4; Vav guanine nucleotide exchange factor 3 (VAV3); breast cancer; cancer; cell migration; guanine nucleotide exchange factor (GEF); phosphotyrosine signaling; protein-protein interaction; receptor tyrosine kinase.
© 2020 Ojala et al.
Conflict of interest statement
Conflict of interest—K. E. has research agreements with Boehringer Ingelheim and Puma Biotechnology and ownership interest in Abomics, Novo Nordisk, Orion, and Roche.
Figures



Similar articles
-
Vav3 mediates receptor protein tyrosine kinase signaling, regulates GTPase activity, modulates cell morphology, and induces cell transformation.Mol Cell Biol. 2000 Dec;20(24):9212-24. doi: 10.1128/MCB.20.24.9212-9224.2000. Mol Cell Biol. 2000. PMID: 11094073 Free PMC article.
-
Vav family proteins couple to diverse cell surface receptors.Mol Cell Biol. 2000 Sep;20(17):6364-73. doi: 10.1128/MCB.20.17.6364-6373.2000. Mol Cell Biol. 2000. PMID: 10938113 Free PMC article.
-
Essential role of Vav family guanine nucleotide exchange factors in EphA receptor-mediated angiogenesis.Mol Cell Biol. 2006 Jul;26(13):4830-42. doi: 10.1128/MCB.02215-05. Mol Cell Biol. 2006. PMID: 16782872 Free PMC article.
-
Involvement of the guanine nucleotide exchange factor Vav3 in central nervous system development and plasticity.Biol Chem. 2017 May 1;398(5-6):663-675. doi: 10.1515/hsz-2016-0275. Biol Chem. 2017. PMID: 28214347 Review.
-
VAV3 in human cancers: Mechanism and clinical implication.Pathol Res Pract. 2023 Aug;248:154681. doi: 10.1016/j.prp.2023.154681. Epub 2023 Jul 13. Pathol Res Pract. 2023. PMID: 37467637 Review.
Cited by
-
From Cell Lines to Patients: Dissecting the Proteomic Landscape of Exosomes in Breast Cancer.Diagnostics (Basel). 2025 Apr 17;15(8):1028. doi: 10.3390/diagnostics15081028. Diagnostics (Basel). 2025. PMID: 40310419 Free PMC article.
-
The role of STAT3/VAV3 in glucolipid metabolism during the development of HFD-induced MAFLD.Int J Biol Sci. 2024 Mar 11;20(6):2027-2043. doi: 10.7150/ijbs.86465. eCollection 2024. Int J Biol Sci. 2024. PMID: 38617550 Free PMC article.
-
Identification of the Novel Tumor Suppressor Role of FOCAD/miR-491-5p to Inhibit Cancer Stemness, Drug Resistance and Metastasis via Regulating RABIF/MMP Signaling in Triple Negative Breast Cancer.Cells. 2021 Sep 24;10(10):2524. doi: 10.3390/cells10102524. Cells. 2021. PMID: 34685504 Free PMC article.
-
Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets.Biology (Basel). 2021 Jan 8;10(1):39. doi: 10.3390/biology10010039. Biology (Basel). 2021. PMID: 33430106 Free PMC article.
-
Trans-activating mutations of the pseudokinase ERBB3.Oncogene. 2024 Jul;43(29):2253-2265. doi: 10.1038/s41388-024-03070-9. Epub 2024 May 28. Oncogene. 2024. PMID: 38806620 Free PMC article.
References
-
- Elenius K., Corfas G., Paul S., Choi C. J., Rio C., Plowman G. D., and Klagsbrun M. (1997) A novel juxtamembrane domain isoform of HER4/ErbB4: isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 272, 26761–26768 10.1074/jbc.272.42.26761 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

