miR2118-dependent U-rich phasiRNA production in rice anther wall development
- PMID: 32561756
- PMCID: PMC7305157
- DOI: 10.1038/s41467-020-16637-3
miR2118-dependent U-rich phasiRNA production in rice anther wall development
Abstract
Reproduction-specific small RNAs are vital regulators of germline development in animals and plants. MicroRNA2118 (miR2118) is conserved in plants and induces the production of phased small interfering RNAs (phasiRNAs). To reveal the biological functions of miR2118, we describe here rice mutants with large deletions of the miR2118 cluster. Our results demonstrate that the loss of miR2118 causes severe male and female sterility in rice, associated with marked morphological and developmental abnormalities in somatic anther wall cells. Small RNA profiling reveals that miR2118-dependent 21-nucleotide (nt) phasiRNAs in the anther wall are U-rich, distinct from the phasiRNAs in germ cells. Furthermore, the miR2118-dependent biogenesis of 21-nt phasiRNAs may involve the Argonaute proteins OsAGO1b/OsAGO1d, which are abundant in anther wall cell layers. Our study highlights the site-specific differences of phasiRNAs between somatic anther wall and germ cells, and demonstrates the significance of miR2118/U-phasiRNA functions in anther wall development and rice reproduction.
Conflict of interest statement
The authors declare no competing interests.
Figures

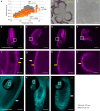
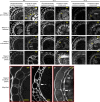
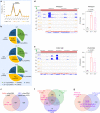
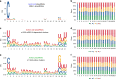

References
-
- Ketting RF. The many faces of RNAi. Dev. Cell. 2011;20:148–161. - PubMed
-
- Ma Z, Zhang X. Actions of plant Argonautes: predictable or unpredictable? Curr. Opin. Plant Biol. 2018;45:59–67. - PubMed
-
- Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

