Genomic signatures of domestication in Old World camels
- PMID: 32561887
- PMCID: PMC7305198
- DOI: 10.1038/s42003-020-1039-5
Genomic signatures of domestication in Old World camels
Abstract
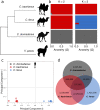
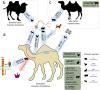
Domestication begins with the selection of animals showing less fear of humans. In most domesticates, selection signals for tameness have been superimposed by intensive breeding for economical or other desirable traits. Old World camels, conversely, have maintained high genetic variation and lack secondary bottlenecks associated with breed development. By re-sequencing multiple genomes from dromedaries, Bactrian camels, and their endangered wild relatives, here we show that positive selection for candidate genes underlying traits collectively referred to as 'domestication syndrome' is consistent with neural crest deficiencies and altered thyroid hormone-based signaling. Comparing our results with other domestic species, we postulate that the core set of domestication genes is considerably smaller than the pan-domestication set - and overlapping genes are likely a result of chance and redundancy. These results, along with the extensive genomic resources provided, are an important contribution to understanding the evolutionary history of camels and the genomic features of their domestication.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Vigne J-D. The origins of animal domestication and husbandry: a major change in the history of humanity and the biosphere. C. R. Biol. 2011;334:171–181. - PubMed
-
- Jensen P. Behavior genetics and the domestication of animals. Annu. Rev. Anim. Biosci. 2014;2:85–104. - PubMed
-
- Crockford, S. J. in Human evolution through developmental change (eds. Minugh-Purvis N. & McNamara K. J.) 122–153 (Johns Hopkins University Press, 2002).
-
- Wilkins AS. Revisiting two hypotheses on the “domestication syndrome” in light of genomic data. Vavilov J. Genet. Breed. 2017;21:435–442.
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

