KDEL receptor is a cell surface receptor that cycles between the plasma membrane and the Golgi via clathrin-mediated transport carriers
- PMID: 32562023
- PMCID: PMC11072833
- DOI: 10.1007/s00018-020-03570-3
KDEL receptor is a cell surface receptor that cycles between the plasma membrane and the Golgi via clathrin-mediated transport carriers
Abstract
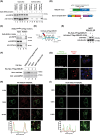
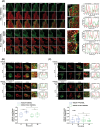
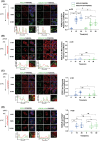
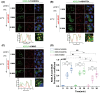
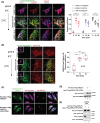
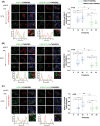
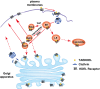
KDEL receptor cycles between the ER and the Golgi to retrieve ER-resident chaperones that get leaked to the secretory pathway during protein export from the ER. Recent studies have shown that a fraction of KDEL receptor may reside in the plasma membrane and function as a putative cell surface receptor. However, the trafficking itinerary and mechanism of cell surface expressed KDEL receptor remains largely unknown. In this study, we used N-terminally Halo-tagged KDEL receptor to investigate its endocytosis from the plasma membrane and trafficking itinerary of the endocytosed receptor through the endolysosomal compartments. Our results indicate that surface-expressed KDEL receptor undergoes highly complex recycling pathways via the Golgi and peri-nuclear recycling endosomes that are positive for Rab11 and Rab14, respectively. Unexpectedly, KDEL receptor appears to preferentially utilize clathrin-mediated endocytic pathway as well as clathrin-dependent transport carriers for export from the trans-Golgi network. Taken together, we suggest that KDEL receptor may be a bona fide cell surface receptor with a complex, yet well-defined trafficking itinerary through the endolysosomal compartments.
Keywords: Caveolae; Clathrin; EEA1; Early endosomes; Golgi; KDEL receptor; Late endosomes; Lysosome; MANF; Membrane trafficking; Rab11; Rab14; Recycling endosomes.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
ACBD3 modulates KDEL receptor interaction with PKA for its trafficking via tubulovesicular carrier.BMC Biol. 2021 Sep 7;19(1):194. doi: 10.1186/s12915-021-01137-7. BMC Biol. 2021. PMID: 34493279 Free PMC article.
-
KDEL Receptor Trafficking to the Plasma Membrane Is Regulated by ACBD3 and Rab4A-GTP.Cells. 2023 Apr 4;12(7):1079. doi: 10.3390/cells12071079. Cells. 2023. PMID: 37048152 Free PMC article.
-
Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling.J Biol Chem. 2003 Feb 28;278(9):7564-72. doi: 10.1074/jbc.M210457200. Epub 2002 Dec 12. J Biol Chem. 2003. PMID: 12482757
-
Regulation of Golgi signaling and trafficking by the KDEL receptor.Histochem Cell Biol. 2013 Oct;140(4):395-405. doi: 10.1007/s00418-013-1130-9. Epub 2013 Jul 20. Histochem Cell Biol. 2013. PMID: 23873287 Review.
-
Bidirectional traffic between the Golgi and the endosomes - machineries and regulation.J Cell Sci. 2016 Nov 1;129(21):3971-3982. doi: 10.1242/jcs.185702. Epub 2016 Oct 10. J Cell Sci. 2016. PMID: 27802132 Review.
Cited by
-
KDEL Receptors: Pathophysiological Functions, Therapeutic Options, and Biotechnological Opportunities.Biomedicines. 2022 May 25;10(6):1234. doi: 10.3390/biomedicines10061234. Biomedicines. 2022. PMID: 35740256 Free PMC article. Review.
-
ACBD3 modulates KDEL receptor interaction with PKA for its trafficking via tubulovesicular carrier.BMC Biol. 2021 Sep 7;19(1):194. doi: 10.1186/s12915-021-01137-7. BMC Biol. 2021. PMID: 34493279 Free PMC article.
-
Ubiquitin-Specific Protease 22 Plays a Key Role in Increasing Extracellular Vesicle Secretion and Regulating Cell Motility of Lung Adenocarcinoma.Adv Sci (Weinh). 2024 Oct;11(38):e2405731. doi: 10.1002/advs.202405731. Epub 2024 Aug 5. Adv Sci (Weinh). 2024. PMID: 39101247 Free PMC article.
-
Directing ricin-based immunotoxins with targeting affibodies and KDEL signal peptide to cancer cells effectively induces apoptosis and tumor suppression.J Nanobiotechnology. 2022 Aug 23;20(1):387. doi: 10.1186/s12951-022-01601-8. J Nanobiotechnology. 2022. PMID: 35999603 Free PMC article.
-
Cerebral Dopamine Neurotrophic Factor (CDNF) Acts as a Trophic Factor Promoting Neuritogenesis in the Dorsal Root Ganglia (DRG) Neurons Through Activation of the PI3K Signaling Pathway.J Neurochem. 2025 Aug;169(8):e70194. doi: 10.1111/jnc.70194. J Neurochem. 2025. PMID: 40827505 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

