A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19
- PMID: 32562705
- PMCID: PMC7299870
- DOI: 10.1016/j.antiviral.2020.104857
A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19
Abstract
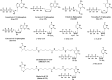
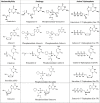
SARS-CoV-2, a member of the coronavirus family, is responsible for the current COVID-19 worldwide pandemic. We previously demonstrated that five nucleotide analogues inhibit the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), including the active triphosphate forms of Sofosbuvir, Alovudine, Zidovudine, Tenofovir alafenamide and Emtricitabine. We report here the evaluation of a library of nucleoside triphosphate analogues with a variety of structural and chemical features as inhibitors of the RdRps of SARS-CoV and SARS-CoV-2. These features include modifications on the sugar (2' or 3' modifications, carbocyclic, acyclic, or dideoxynucleotides) or on the base. The goal is to identify nucleotide analogues that not only terminate RNA synthesis catalyzed by these coronavirus RdRps, but also have the potential to resist the viruses' exonuclease activity. We examined these nucleotide analogues for their ability to be incorporated by the RdRps in the polymerase reaction and to prevent further incorporation. While all 11 molecules tested displayed incorporation, 6 exhibited immediate termination of the polymerase reaction (triphosphates of Carbovir, Ganciclovir, Stavudine and Entecavir; 3'-OMe-UTP and Biotin-16-dUTP), 2 showed delayed termination (Cidofovir diphosphate and 2'-OMe-UTP), and 3 did not terminate the polymerase reaction (2'-F-dUTP, 2'-NH2-dUTP and Desthiobiotin-16-UTP). The coronaviruses possess an exonuclease that apparently requires a 2'-OH at the 3'-terminus of the growing RNA strand for proofreading. In this study, all nucleoside triphosphate analogues evaluated form Watson-Crick-like base pairs. The nucleotide analogues demonstrating termination either lack a 2'-OH, have a blocked 2'-OH, or show delayed termination. Thus, these nucleotide analogues are of interest for further investigation to evaluate whether they can evade the viral exonuclease activity. Prodrugs of five of these nucleotide analogues (Cidofovir, Abacavir, Valganciclovir/Ganciclovir, Stavudine and Entecavir) are FDA-approved medications for treatment of other viral infections, and their safety profiles are well established. After demonstrating potency in inhibiting viral replication in cell culture, candidate molecules can be rapidly evaluated as potential therapies for COVID-19.
Keywords: COVID-19; Exonuclease; Nucleotide analogues; RNA-Dependent RNA polymerase; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

