Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated With Viral Antagonism of STAT1 Phosphorylation
- PMID: 32563187
- PMCID: PMC7337793
- DOI: 10.1093/infdis/jiaa356
Attenuated Interferon and Proinflammatory Response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated With Viral Antagonism of STAT1 Phosphorylation
Abstract
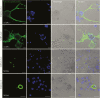
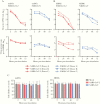
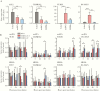
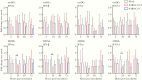
Clinical manifestations of coronavirus disease 2019 (COVID-19) vary from asymptomatic virus shedding, nonspecific pharyngitis, to pneumonia with silent hypoxia and respiratory failure. Dendritic cells and macrophages are sentinel cells for innate and adaptive immunity that affect the pathogenesis of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). The interplay between SARS-CoV-2 and these cell types remains unknown. We investigated infection and host responses of monocyte-derived dendritic cells (moDCs) and macrophages (MDMs) infected by SARS-CoV-2. MoDCs and MDMs were permissive to SARS-CoV-2 infection and protein expression but did not support productive virus replication. Importantly, SARS-CoV-2 launched an attenuated interferon response in both cell types and triggered significant proinflammatory cytokine/chemokine expression in MDMs but not moDCs. Investigations suggested that this attenuated immune response to SARS-CoV-2 in moDCs was associated with viral antagonism of STAT1 phosphorylation. These findings may explain the mild and insidious course of COVID-19 until late deterioration.
Keywords: COVID-19; MDMs; SARS-CoV-2; coronavirus; dendritic cells; macrophages; moDCs.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures


References
-
- World Health Organization. Coronavirus disease (COVID-19) situation report–108 https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed 23 June 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

