The promise of single-cell mechanophenotyping for clinical applications
- PMID: 32566069
- PMCID: PMC7286698
- DOI: 10.1063/5.0010800
The promise of single-cell mechanophenotyping for clinical applications
Abstract
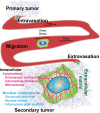
Cancer is the second leading cause of death worldwide. Despite the immense research focused in this area, one is still not able to predict disease trajectory. To overcome shortcomings in cancer disease study and monitoring, we describe an exciting research direction: cellular mechanophenotyping. Cancer cells must overcome many challenges involving external forces from neighboring cells, the extracellular matrix, and the vasculature to survive and thrive. Identifying and understanding their mechanical behavior in response to these forces would advance our understanding of cancer. Moreover, used alongside traditional methods of immunostaining and genetic analysis, mechanophenotyping could provide a comprehensive view of a heterogeneous tumor. In this perspective, we focus on new technologies that enable single-cell mechanophenotyping. Single-cell analysis is vitally important, as mechanical stimuli from the environment may obscure the inherent mechanical properties of a cell that can change over time. Moreover, bulk studies mask the heterogeneity in mechanical properties of single cells, especially those rare subpopulations that aggressively lead to cancer progression or therapeutic resistance. The technologies on which we focus include atomic force microscopy, suspended microchannel resonators, hydrodynamic and optical stretching, and mechano-node pore sensing. These technologies are poised to contribute to our understanding of disease progression as well as present clinical opportunities.
Copyright © 2020 Author(s).
Figures

Similar articles
-
The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells.Rep Prog Phys. 2019 Jun;82(6):064602. doi: 10.1088/1361-6633/ab1628. Epub 2019 Apr 4. Rep Prog Phys. 2019. PMID: 30947151 Review.
-
The fundamental role of mechanical properties in the progression of cancer disease and inflammation.Rep Prog Phys. 2014 Jul;77(7):076602. doi: 10.1088/0034-4885/77/7/076602. Epub 2014 Jul 9. Rep Prog Phys. 2014. PMID: 25006689 Review.
-
Molecular Tension Probes for Imaging Forces at the Cell Surface.Acc Chem Res. 2017 Dec 19;50(12):2915-2924. doi: 10.1021/acs.accounts.7b00305. Epub 2017 Nov 21. Acc Chem Res. 2017. PMID: 29160067 Free PMC article. Review.
-
A combined experimental and theoretical approach towards mechanophenotyping of biological cells using a constricted microchannel.Lab Chip. 2017 Oct 25;17(21):3704-3716. doi: 10.1039/c7lc00599g. Lab Chip. 2017. PMID: 28983550
-
Right care, first time: a highly personalised and measurement-based care model to manage youth mental health.Med J Aust. 2019 Nov;211 Suppl 9:S3-S46. doi: 10.5694/mja2.50383. Med J Aust. 2019. PMID: 31679171
Cited by
-
Single-Cell Mechanophenotyping in Microfluidics to Evaluate Behavior of U87 Glioma Cells.Micromachines (Basel). 2020 Sep 11;11(9):845. doi: 10.3390/mi11090845. Micromachines (Basel). 2020. PMID: 32932941 Free PMC article.
-
Mechanical phenotyping reveals unique biomechanical responses in retinoic acid-resistant acute promyelocytic leukemia.iScience. 2022 Jan 15;25(2):103772. doi: 10.1016/j.isci.2022.103772. eCollection 2022 Feb 18. iScience. 2022. PMID: 35141508 Free PMC article.
-
Mechanical Measurements of Cells Using AFM: 3D or 2D Physics?Front Bioeng Biotechnol. 2020 Nov 19;8:605153. doi: 10.3389/fbioe.2020.605153. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33330437 Free PMC article. No abstract available.
-
Bacteria Residing at Root Canals Can Induce Cell Proliferation and Alter the Mechanical Properties of Gingival and Cancer Cells.Int J Mol Sci. 2020 Oct 24;21(21):7914. doi: 10.3390/ijms21217914. Int J Mol Sci. 2020. PMID: 33114460 Free PMC article.
-
Molecular mechanocytometry using tension-activated cell tagging.Nat Methods. 2023 Nov;20(11):1666-1671. doi: 10.1038/s41592-023-02030-7. Epub 2023 Oct 5. Nat Methods. 2023. PMID: 37798479 Free PMC article.
References
-
- Fejerman L., Stern M. C., John E. M., Torres-Mejia G., Hines L. M., Wolff R. K., Baumgartner K. B., Giuliano A. R., Ziv E., Perez-Stable E. J., and Slattery M. L., “Interaction between common breast cancer susceptibility variants, genetic ancestry, and nongenetic risk factors in Hispanic women,” Cancer Epidemiol. Biomarkers Prev. 24(11), 1731–1738 (2015). 10.1158/1055-9965.EPI-15-0392 - DOI - PMC - PubMed
-
- Huyghe J. R., Bien S. A., Harrison T. A., Kang H. M., Chen S., Schmit S. L., Conti D. V., Qu C., Jeon J., Edlund C. K., Greenside P., Wainberg M., Schumacher F. R., Smith J. D., Levine D. M., Nelson S. C., Sinnott-Armstrong N. A., Albanes D., Alonso M. H., Anderson K., Arnau-Collell C., Arndt V., Bamia C., Banbury B. L., Baron J. A., Berndt S. I., Bezieau S., Bishop D. T., Boehm J., Boeing H., Brenner H., Brezina S., Buch S., Buchanan D. D., Burnett-Hartman A., Butterbach K., Caan B. J., Campbell P. T., Carlson C. S., Castellvi-Bel S., Chan A. T., Chang-Claude J., Chanock S. J., Chirlaque M. D., Cho S. H., Connolly C. M., Cross A. J., Cuk K., Curtis K. R., de la Chapelle A., Doheny K. F., Duggan D., Easton D. F., Elias S. G., Elliott F., English D. R., Feskens E. J. M., Figueiredo J. C., Fischer R., FitzGerald L. M., Forman D., Gala M., Gallinger S., Gauderman W. J., Giles G. G., Gillanders E., Gong J., Goodman P. J., Grady W. M., Grove J. S., Gsur A., Gunter M. J., Haile R. W., Hampe J., Hampel H., Harlid S., Hayes R. B., Hofer P., Hoffmeister M., Hopper J. L., Hsu W. L., Huang W. Y., Hudson T. J., Hunter D. J., Ibanez-Sanz G., Idos G. E., Ingersoll R., Jackson R. D., Jacobs E. J., Jenkins M. A., Joshi A. D., Joshu C. E., Keku T. O., Key T. J., Kim H. R., Kobayashi E., Kolonel L. N., Kooperberg C., Kuhn T., Kury S., Kweon S. S., Larsson S. C., Laurie C. A., Le Marchand L., Leal S. M., Lee S. C., Lejbkowicz F., Lemire M., Li C. I., Li L., Lieb W., Lin Y., Lindblom A., Lindor N. M., Ling H., Louie T. L., Mannisto S., Markowitz S. D., Martin V., Masala G., McNeil C. E., Melas M., Milne R. L., Moreno L., Murphy N., Myte R., Naccarati A., Newcomb P. A., Offit K., Ogino S., Onland-Moret N. C., Pardini B., Parfrey P. S., Pearlman R., Perduca V., Pharoah P. D. P., Pinchev M., Platz E. A., Prentice R. L., Pugh E., Raskin L., Rennert G., Rennert H. S., Riboli E., Rodriguez-Barranco M., Romm J., Sakoda L. C., Schafmayer C., Schoen R. E., Seminara D., Shah M., Shelford T., Shin M. H., Shulman K., Sieri S., Slattery M. L., Southey M. C., Stadler Z. K., Stegmaier C., Su Y. R., Tangen C. M., Thibodeau S. N., Thomas D. C., Thomas S. S., Toland A. E., Trichopoulou A., Ulrich C. M., Van Den Berg D. J., van Duijnhoven F. J. B., Van Guelpen B., van Kranen H., Vijai J., Visvanathan K., Vodicka P., Vodickova L., Vymetalkova V., Weigl K., Weinstein S. J., White E., Win A. K., Wolf C. R., Wolk A., Woods M. O., Wu A. H., Zaidi S. H., Zanke B. W., Zhang Q., Zheng W., Scacheri P. C., Potter J. D., Bassik M. C., Kundaje A., Casey G., Moreno V., Abecasis G. R., Nickerson D. A., Gruber S. B., Hsu L., and Peters U., “Discovery of common and rare genetic risk variants for colorectal cancer,” Nat. Genet. 51(1), 76–87 (2019). 10.1038/s41588-018-0286-6 - DOI - PMC - PubMed
