DiscoSnp-RAD: de novo detection of small variants for RAD-Seq population genomics
- PMID: 32566401
- PMCID: PMC7293188
- DOI: 10.7717/peerj.9291
DiscoSnp-RAD: de novo detection of small variants for RAD-Seq population genomics
Abstract
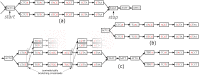
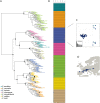
Restriction site Associated DNA Sequencing (RAD-Seq) is a technique characterized by the sequencing of specific loci along the genome that is widely employed in the field of evolutionary biology since it allows to exploit variants (mainly Single Nucleotide Polymorphism-SNPs) information from entire populations at a reduced cost. Common RAD dedicated tools, such as STACKS or IPyRAD, are based on all-vs-all read alignments, which require consequent time and computing resources. We present an original method, DiscoSnp-RAD, that avoids this pitfall since variants are detected by exploiting specific parts of the assembly graph built from the reads, hence preventing all-vs-all read alignments. We tested the implementation on simulated datasets of increasing size, up to 1,000 samples, and on real RAD-Seq data from 259 specimens of Chiastocheta flies, morphologically assigned to seven species. All individuals were successfully assigned to their species using both STRUCTURE and Maximum Likelihood phylogenetic reconstruction. Moreover, identified variants succeeded to reveal a within-species genetic structure linked to the geographic distribution. Furthermore, our results show that DiscoSnp-RAD is significantly faster than state-of-the-art tools. The overall results show that DiscoSnp-RAD is suitable to identify variants from RAD-Seq data, it does not require time-consuming parameterization steps and it stands out from other tools due to its completely different principle, making it substantially faster, in particular on large datasets.
Keywords: Deletions; Insertions; RAD-seq; Reference-free; SNPs; Variants.
© 2020 Gauthier et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

