Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment
- PMID: 32567735
- PMCID: PMC7396846
- DOI: 10.15252/embj.2019103790
Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment
Abstract
Tumour-associated microglia/macrophages (TAM) are the most numerous non-neoplastic populations in the tumour microenvironment in glioblastoma multiforme (GBM), the most common malignant brain tumour in adulthood. The mTOR pathway, an important regulator of cell survival/proliferation, is upregulated in GBM, but little is known about the potential role of this pathway in TAM. Here, we show that GBM-initiating cells induce mTOR signalling in the microglia but not bone marrow-derived macrophages in both in vitro and in vivo GBM mouse models. mTOR-dependent regulation of STAT3 and NF-κB activity promotes an immunosuppressive microglial phenotype. This hinders effector T-cell infiltration, proliferation and immune reactivity, thereby contributing to tumour immune evasion and promoting tumour growth in mouse models. The translational value of our results is demonstrated in whole transcriptome datasets of human GBM and in a novel in vitro model, whereby expanded-potential stem cells (EPSC)-derived microglia-like cells are conditioned by syngeneic patient-derived GBM-initiating cells. These results raise the possibility that microglia could be the primary target of mTOR inhibition, rather than the intrinsic tumour cells in GBM.
Keywords: TAM; mTOR; T cells; glioblastoma; microglia.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
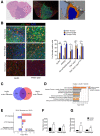
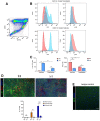
- A
Representative images of a PDGFB tumour: H&E on the right to identify the tumour, an adjacent section stained for Iba1 (red), p‐S6 (green) and DAPI (blue) at the centre and the region selection for quantification on the left (non‐tumour in light blue, tumour edge in orange and tumour core in dark blue).
- B
Representative images of core, edge and adjacent non‐tumour brain tissue for Iba1, p‐S6 and DAPI staining in Pten −/− p53 −/− (right) and GL261 (left) tumours. Percentage of Iba1+ cells co‐expressing p‐S6 in the three defined regions. Five high‐grade glioma models were analysed—GL261 (n = 3), PDGFB (n = 3), Ntv‐a;PDGFB+shp53 (n = 2), Pten −/− p53 −/− (n = 3) and Pten −/− p53 −/− Idh1 mut (n = 3) (mean ± SEM; two‐way ANOVA Tukey test).
- C
Venn diagram identifying significantly deregulated genes in healthy versus GL261 TAM‐MG and/or TAM‐BMDM (GSE68376 dataset).
- D
Top‐most deregulated canonical pathways in GL261 TAM‐MG, as identified by the IPA software. Threshold indicates P ≤ 0.05. Z‐score indicates the orientation of the deregulation. Ratio indicates the number of deregulated genes in the pathway.
- E
mTOR‐related deregulated canonical pathways in GL261 TAM‐MG and TAM‐BMDM, as identified by the IPA software.
- F, G
Expression levels of (F) Rps6 and (G) Eif4ebp1 in healthy versus GL261 microglia and macrophage (n = 3; mean ± SEM, likelihood ratio test in edgeR).
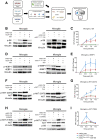
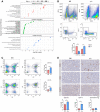
- A
Schematic of the in vitro model whereby microglia and BMDM were pretreated with Torin, LY294002 as indicated and stimulated with mGL261, mGICPten−/−;p53−/−‐CM or mNSC‐CM.
- B–I
Signalling was analysed in microglia by immunoblotting of whole cell lysates collected at 4 h (B, D and F) and 0.5 h (H) and normalised against non‐phosphorylated protein and vinculin analysed on the same blot. Flow cytometry analysis was carried out in microglia for (C) p‐S6 S240/244; (E) p‐4EBP1 T37/46; (G) p‐AKT S473; and (I) p‐AKT T308. Each treatment (mNSC‐CM, Pten −/− p53 −/− mGIC‐CM, Pten −/− p53 −/− mGIC‐CM+Torin, mGIC‐CM+LY) was normalised to unconditioned control (n = 3; mean ± SEM, two‐way ANOVA Tukey test). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 comparing mGICPten−/−;p53−/−‐CM versus mGICPten−/−;p53−/−‐CM +inhibitor.
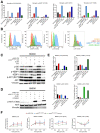
- A
Quantification of immunoblotting analysis from conditioned microglia whole cell lysates at 4 h incubation for p‐S6, p‐4EBP1 and p‐AKT S473 and at 0.5 h incubation for p‐AKT T308. Data normalised to non‐phosphorylated protein and to vinculin.
- B
Representative flow cytometry plots of microglia under different culture condition at 4 h incubation for p‐S6, p‐4EBP1 and p‐AKT S473 and at 0.5 h incubation for p‐AKT T308.
- C–E
Signalling was analysed in conditioned BMDM by immunoblotting of whole cell lysate collected at 4 h for p‐S6, p‐4EBP1 and p‐AKT S473 (C) and at 0.5 h incubation for p‐AKT T308 (D), which was quantified by normalisation with non‐phosphorylated protein and vinculin (E).
- F
Flow cytometry analysis was carried out in BMDM for p‐S6, p‐4EBP1, p‐AKT S473 and p‐AKT T308. Each treatment was normalised to unconditioned control (n = 3; mean ± SEM; two‐way ANOVA Tukey test).
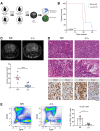
Schematic of the generation and analysis of the Cx3cr1‐Cre;Rheb1‐loxp GL261 model.
Survival analysis for Cx3cr1‐Rheb1 Δ/Δ (n = 7) and Rheb1 fl/fl (n = 7) mice. Chi‐square test.
Representative images and quantification of the tumour volume with MRI of Rheb1 fl/fl (n = 8) compared to Cx3cr1‐Rheb1 Δ/Δ (n = 8) mice (mean ± SEM; unpaired parametric t‐test).
H&E staining showing representative histological features (overview and high magnification of microvascular proliferation) of Rheb1 fl/fl and Cx3cr1‐Rheb1 Δ/Δ GL261 tumours. Scale bar is 125 μm (top, H&E) and 80 μm (all other images)
Percentage of GFP+ CD45− GL261 tumour cells in Cx3cr1‐Rheb1 Δ/Δ (n = 5) and Rheb1 fl/fl (n = 7) tumours, with representative FACS plot (mean ± SEM; unpaired parametric t‐test).
Representative flow cytometry plot for the expression of CD45 and CD11b in GL261 tumour, with CD45high CD11b+ TAM‐BMDM (top gate) and CD45low CD11b+ TAM‐MG (lower gate).
Representative flow cytometry plot of YFP (left) and p‐S6 (right) levels in TAM‐BMDM (top) and TAM‐MG (bottom) in Cx3cr1‐Rheb1 Δ/Δ (blue) versus Rheb1 fl/fl (red) GL261 tumours.
Percentage of TAM‐MG and TAM‐BMDM expressing YFP in Cx3cr1‐Rheb1 Δ/Δ (n = 6) and Rheb1 fl/fl (n = 6) mice. MFI levels of p‐S6 in P2RY12+ CD49d− TAM‐MG and P2RY12− CD49d+ TAM‐BMDM in Rheb1 fl/fl (n = 6) compared to Cx3cr1‐Rheb1 Δ/Δ (n = 6) GL261 tumours (mean ± SEM; two‐way ANOVA Tukey test).
Staining for Iba1, p‐S6 and DAPI in Rheb1 fl/fl (n = 3) and Cx3cr1‐Rheb1 Δ/Δ (n = 2) tumour tissue (top) and percentage of Iba1+ cells co‐expressing p‐S6 in the three defined regions (bottom) (mean ± SEM; two‐way ANOVA Tukey test). Scale bar is 100 μm.
Isotype control stain demonstrating quenching of fluorescence from reporter gene (scale bar 50 μm).
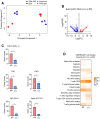
Principal component analysis of bulk (triangle) and TAM‐MG (circle) RNA‐Seq samples from Rheb1 fl/fl (red) and Cx3cr1‐Rheb1 Δ/Δ (blue) Gl261 tumours.
Volcano plot of differentially expressed genes between Rheb1 fl/fl (n = 3) and Cx3cr1‐Rheb1 Δ/Δ (n = 3) GL261 tumours. Red and blue points mark genes with significantly increased or decreased expression, respectively (FDR ≤ 0.05). The x‐axis shows expression log fold changes (FC), and the y‐axis shows the –log of the false discovery rate (FDR).
Expression levels (TPM) of Csf1r, Csf1, Aif1 (Iba1), Itgam (Cd11b) and Cd274 (PD‐L1) in Cx3cr1‐Rheb1 Δ/Δ (n = 3) versus Rheb1 fl/fl (n = 3) (mean ± SEM, likelihood ratio test in edgeR). *FDR ≤ 0.05, ***FDR ≤ 0.001.
CIBERSORT cell fractions calculated from the TPM of Cx3cr1‐Rheb1 Δ/Δ (n = 3) versus Rheb1 fl/fl (n = 3; mean ± SEM; two‐way ANOVA Tukey test). *P ≤ 0.05.
Significantly deregulated canonical pathways in Cx3cr1‐Rheb1 Δ/Δ versus Rheb1 fl/fl GL261 tumours, as identified by the IPA software from differentially regulated genes and divided into three categories: adaptive immunity, cytokine signalling and innate immunity. Size of bubbles is indicative of ratio of differentially regulated genes. The x‐axis represents −log10 (P‐value) with a threshold of P ≤ 0.05 applied.
Levels of TAM‐MG (P2RY12+ CD49d−) and TAM‐BMDM (P2RY12− CD49d+) gated from CD45+ population in GL261 tumours from Rheb1 fl/fl (n = 4) and Cx3cr1‐Rheb1 Δ/Δ (n = 5) mice by flow cytometry (representative flow cytometry plots on top and quantification at the bottom; mean ± SEM; two‐way ANOVA Tukey test).
Percentage of CD8 CTL (top) and CD4 Th (CD4) cells in GL261 tumours from Rheb1 fl/fl (n = 4) and Cx3cr1‐Rheb1 Δ/Δ (n = 5) mice assessed by flow cytometry (representative flow cytometry plots, left and quantification, right; mean ± SEM; unpaired t‐test).
Tumour tissues from Rheb1 fl/fl (n = 4) and Cx3cr1‐Rheb1 Δ/Δ (n = 4) were stained for CD8, CD4 and FoxP3. DAB staining was quantified as % of positive cells using Definiens software. Representative images of the stained tissues are shown (mean ± SEM; unpaired parametric t‐test). Scale bar is 100 μm.

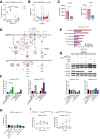
- A
Schematic of the analysis of bulk GL261 RNA‐Seq data from Cx3cr1‐Rheb1 Δ/Δ (n = 3) and Rheb1 fl/fl (n = 3) mice. ssGSEA enrichment scores for signalling pathway signatures and CIBERSORT cell fractions were calculated for each sample. Pearson correlation score was then calculated comparing signalling pathway enrichment scores and CIBERSORT cell fractions. Finally, a differential correlation analysis was run between the Cx3cr1‐Rheb1 Δ/Δ (n = 3) and Rheb1 fl/fl (n = 3) samples.
- B
Heatmap of the differential correlation analysis between CIBERSORT cell fractions and signatures enrichment scores (ssGSEA) for lymphocytes in the Cx3cr1‐Rheb1 Δ/Δ (n = 3) vs. Rheb1 fl/fl (n = 3) GL261 tumours.
- C, D
CD4 Th cells (C) and CD8 CTL cells (D) expression of Ki67, perforin, granzyme b and IFNγ, in Cx3cr1‐Rheb1 fl/fl (n = 5) and Rheb1 Δ/Δ (n = 5) GL261 tumours. Representative FACS plots show control samples (tumour‐derived cells stimulated in culture in the absence of protein inhibitor cocktail—in orange) compared to stimulated cells derived from Rheb1 fl/fl (fl/fl, red) and Cx3cr1‐Rheb1 Δ/Δ (Δ/Δ, blue) GL261 tumours (cells stimulated in culture with protein inhibitor cocktail) (mean ± SEM; two‐way ANOVA Tukey test).
- E, F
Expression of CD44 (E) and CD62L (F) by CD4 Th and CD8 CTL in Cx3cr1‐Rheb1 fl/fl (n = 5; fl/fl, red) and Rheb1 Δ/Δ (n = 5; Δ/Δ, blue) GL261 tumours with representative FACS plots show control samples (mean ± SEM; two‐way ANOVA Tukey test).
ssGSEA enrichment scores from TAM‐MG and TAM‐BMDM signature in Rheb1 fl/fl (n = 2) and Cx3cr1‐Rheb1 Δ/Δ (n = 3) RNA‐Seq from TAM‐MG of GL261 tumours (mean ± SEM; two‐way ANOVA Tukey tests).
Volcano plot of differential expression analysis of genes between Rheb1 fl/fl (n = 2) and Cx3cr1‐Rheb1 Δ/Δ (n = 3) TAM‐MG. Red and blue points mark genes with significantly increased or decreased expression, respectively (P ≤ 0.02). The x‐axis shows expression log fold changes (FC), and the y‐axis shows the –log of the false discovery rate (P‐value).
Expression levels of Isg15 and Usp18 in Cx3cr1‐Rheb1 Δ/Δ (n = 3) versus Rheb1 fl/fl TAM‐MG (n = 2) as calculated from the DEG analysis (mean ± SEM, likelihood ratio test in edgeR).
Schematic representation of the NF‐ĸB pathway with genes deregulated in Cx3cr1‐Rheb1 Δ/Δ versus Rheb1 fl/fl TAM‐MG. Pink circles indicate deregulated expression of the protein, which when coloured in pink upregulated.
Deregulated canonical pathways in GL261 TAM‐MG and TAM‐BMDM, as identified by the IPA software between Cx3cr1‐Rheb1 Δ/Δ and Rheb1 fl/fl TAM‐MG.
Quantification of immunoblotting analysis from conditioned microglia whole cell lysates at 4 h incubation for p‐NF‐κB (p‐P65) (n = 3) and p‐STAT3 (n = 1), normalised to non‐phosphorylated protein and vinculin (mean ± SEM; one‐way ANOVA Tukey tests).
Signalling was analysed in conditioned BMDM by immunoblotting of whole cell lysates collected at 4 h for p‐NF‐κB (p‐P65) and p‐STAT3, which was normalised to non‐phosphorylated protein and vinculin.
Quantification of nuclear translocation of NF‐κB (P65) from immunofluorescence staining. Unit represent the % of voxels in the NF‐κB channel colocalised with DAPI (n = 3; mean ± SEM; one‐way ANOVA Tukey tests).
Flow cytometry analysis of p‐STAT3 in conditioned microglia and BMDM (n = 3; mean ± SEM; two‐way ANOVA Tukey tests).
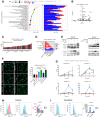
- A
Significantly deregulated canonical pathways in GL261 Rheb1 fl/fl versus Rheb1 Δ/Δ TAM‐MG, as identified by the IPA software from differentially regulated genes. Size of bubbles is indicative of ratio of differentially regulated genes. The x‐axis represents −log10 (P‐value) with a threshold of P ≤ 0.05 applied. On the right, number of up and down regulated genes in each identified pathway is indicated in blue and red respectively.
- B
Frequency of occurrence of differentially regulated genes across significantly deregulated canonical pathways compared to the log fold change (Log(FC)).
- C
Upstream regulators as identified by the IPA software from differentially regulated genes.
- D
Heatmap of Pearson correlation analysis between ssGSEA signatures enrichment scores in the Rheb1 fl/fl versus Cx3cr1‐Rheb1 Δ/Δ GL261 tumours.
- E
Microglia were pretreated with Torin or medium as indicated and then stimulated with mNSC‐CM, mGICPten−/−;p53−/−‐CM or GL261‐CM. p‐NF‐κB (p‐P65) and p‐STAT3 were analysed by immunoblotting of whole cell lysates collected at 4 h.
- F
Immunofluorescence images of NF‐κB (P65) in conditioned microglia. Quantification of nuclear translocation of NF‐κB from staining. Units represent the % of voxels in the NF‐κB channel colocalised with DAPI (n = 3; mean ± SEM; one‐way ANOVA Tukey test—right). Scale bar is 50 μm.
- G
Production of Il‐12 (Il‐12p40), Tnf, Il6 and Il10 by conditioned microglia was determined by qPCR. Each treatment was normalised to housekeeping gene and the unconditioned control (n = 3; mean ± SEM; two‐way ANOVA Tukey test).
- H, I
MFI levels of pSTAT3 in TAM‐MG (CD45+ P2RY12+ CD49d−) (H) and p‐NF‐κB (p‐P65) in TAM‐BMDM (CD45+ P2RY12− CD49d+) (I) from Rheb1 fl/fl (fl/fl, blue, n = 4) and Cx3cr1‐Rheb1 Δ/Δ (Δ/Δ, red, n = 5) GL261 tumours. Representative FACS plots are displayed for each cell type (mean ± SEM, two‐way ANOVA Tukey test).
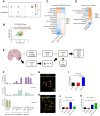
- A
Correlation between ssGSEA enrichment scores for the mTOR signature versus TAM‐MG or TAM‐BMDM signatures in TCGA‐GBM transcriptomic data. Comparison carried out on all IDH‐wild‐type samples and in a subgroup‐specific manner according to Wang's classifier. Size of circle is indicative of R‐square value, and bold outline represents a P ≤ 0.05.
- B
Separation of IDH‐wild‐type GBM samples between those displaying high mTOR and microglia enrichment (+ve correlation, group 1 in orange) and those without this signature (group 2 in green).
- C
CIBERSORT cell fractions calculated from the TPM of TCGA IDH‐wild‐type GBM samples from group 1 compared to those from group 2.
- D
Heatmap of Pearson correlation analysis comparing ssGSEA enrichment scores of TAM‐MG and TAM‐BMDM signatures versus signalling pathway signatures, calculated from the TPM of IDH‐wild‐type GBM samples with the signature from group 1.
- E
Schematic of the experimental setup to derive patient‐matched EPSC and GIC lines.
- F
Staining by flow cytometry of EPSC, YS‐EBs and matched‐iMGL with indicated markers (n = 3; mean ± SEM).
- G, H
Mature iMGL were immunostained for (G) PU1 and CD11b and (H) Iba1 and TREM2. Scale bar 20 μm.
- I–K
(I) P‐S6, (J) p‐STAT3 and (K) p‐NF‐κB (p‐P65) were analysed by flow cytometry in iMGL, pretreated with Torin, as indicated and then stimulated with matched patient EPSC‐ or hGIC‐CM. (n = 3; mean ± SEM, two‐way ANOVA Tukey test).
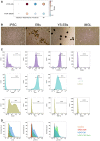
Correlation between ssGSEA enrichment scores for the mTOR signature versus TAM‐MG or TAM‐BMDM signatures in Gravendeel‐GBM transcriptomic data. Comparison carried out on all IDH‐wild‐type samples and in a subgroup‐specific manner according to Wang's classifier. Size of circle is indicative of R‐square value and bold outline represents a P ≤ 0.05.
Representative images of differentiation protocol of EPSC into EBS, YS‐EBs and iMGL. Scale bar is 50 μm.
Representative flow cytometry plots for changes in expression of surface markers during differentiation of EPSC to iMGL. Histogram of expression of SOX2 (stem cell marker), PU.1 (early yolk sac marker), P2RY12 (microglia‐specific marker) and CD49d (macrophage specific marker) by EPSC, YS‐EBs and iMGL are shown.
Representative flow cytometry plots of expression of p‐S6, p‐STAT3 and p‐NF‐κB (p‐P65) in iMGL following 4 h of incubation with different culture conditions.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- NC/L000954/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- P30 CA016672/CA/NCI NIH HHS/United States
- G0800020/MRC_/Medical Research Council/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- MR/N000528/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

