Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19
- PMID: 32567979
- PMCID: PMC7332868
- DOI: 10.1080/07391102.2020.1778535
Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19
Abstract
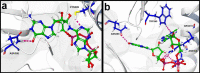
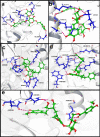
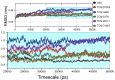
The recent pandemic caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) calls the whole world into a medical emergency. For tackling Coronavirus Disease 2019 (COVID-19), researchers from around the world are swiftly working on designing and identifying inhibitors against all possible viral key protein targets. One of the attractive drug targets is guanine-N7 methyltransferase which plays the main role in capping the 5'-ends of viral genomic RNA and sub genomic RNAs, to escape the host's innate immunity. We performed homology modeling and molecular dynamic (MD) simulation, in order to understand the molecular architecture of Guanosine-P3-Adenosine-5',5'-Triphosphate (G3A) binding with C-terminal N7-MTase domain of nsp14 from SARS-CoV-2. The residue Asn388 is highly conserved in present both in N7-MTase from SARS-CoV and SARS-CoV-2 and displays a unique function in G3A binding. For an in-depth understanding of these substrate specificities, we tried to screen and identify inhibitors from the Traditional Chinese Medicine (TCM) database. The combination of several computational approaches, including screening, MM/GBSA, MD simulations, and PCA calculations, provides the screened compounds that readily interact with the G3A binding site of homology modeled N7-MTase domain. Compounds from this screening will have strong potency towards inhibiting the substrate-binding and efficiently hinder the viral 5'-end RNA capping mechanism. We strongly believe the final compounds can become COVID-19 therapeutics, with huge international support.[Formula: see text]The focus of this study is to screen for antiviral inhibitors blocking guanine-N7 methyltransferase (N7-MTase), one of the key drug targets involved in the first methylation step of the SARS-CoV-2 RNA capping mechanism. Compounds binding the substrate-binding site can interfere with enzyme catalysis and impede 5'-end cap formation, which is crucial to mimic host RNA and evade host cellular immune responses. Therefore, our study proposes the top hit compounds from the Traditional Chinese Medicine (TCM) database using a combination of several computational approaches.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Methyltransferase; N7-MTase; RNA capping; SARS-CoV-2; TCM; ensemble sampling; molecular dynamics; natural products; nsp14.
Figures


Similar articles
-
Opportunities and Challenges in Targeting the Proofreading Activity of SARS-CoV-2 Polymerase Complex.Molecules. 2022 May 3;27(9):2918. doi: 10.3390/molecules27092918. Molecules. 2022. PMID: 35566268 Free PMC article. Review.
-
Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase.Eur J Med Chem. 2020 Sep 1;201:112557. doi: 10.1016/j.ejmech.2020.112557. Epub 2020 Jun 12. Eur J Med Chem. 2020. PMID: 32563813 Free PMC article.
-
In silico evaluation of S-adenosyl-L-homocysteine analogs as inhibitors of nsp14-viral cap N7 methyltranferase and PLpro of SARS-CoV-2: synthesis, molecular docking, physicochemical data, ADMET and molecular dynamics simulations studies.J Biomol Struct Dyn. 2025 Apr;43(7):3258-3275. doi: 10.1080/07391102.2023.2297005. Epub 2023 Dec 26. J Biomol Struct Dyn. 2025. PMID: 38147408
-
Mutagenesis of S-Adenosyl-l-Methionine-Binding Residues in Coronavirus nsp14 N7-Methyltransferase Demonstrates Differing Requirements for Genome Translation and Resistance to Innate Immunity.J Virol. 2016 Jul 27;90(16):7248-7256. doi: 10.1128/JVI.00542-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252528 Free PMC article.
-
Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target.J Med Virol. 2021 Jul;93(7):4258-4264. doi: 10.1002/jmv.27009. Epub 2021 Apr 23. J Med Virol. 2021. PMID: 33837972 Free PMC article. Review.
Cited by
-
Biomolecular interactions with nanoparticles: applications for coronavirus disease 2019.Curr Opin Colloid Interface Sci. 2021 Aug;54:101461. doi: 10.1016/j.cocis.2021.101461. Epub 2021 Apr 23. Curr Opin Colloid Interface Sci. 2021. PMID: 33907504 Free PMC article. Review.
-
A universal fluorescence polarization high throughput screening assay to target the SAM-binding sites of SARS-CoV-2 and other viral methyltransferases.Emerg Microbes Infect. 2023 Dec;12(1):2204164. doi: 10.1080/22221751.2023.2204164. Emerg Microbes Infect. 2023. PMID: 37060263 Free PMC article.
-
Novel Drug Design for Treatment of COVID-19: A Systematic Review of Preclinical Studies.Can J Infect Dis Med Microbiol. 2022 Sep 25;2022:2044282. doi: 10.1155/2022/2044282. eCollection 2022. Can J Infect Dis Med Microbiol. 2022. PMID: 36199815 Free PMC article. Review.
-
Opportunities and Challenges in Targeting the Proofreading Activity of SARS-CoV-2 Polymerase Complex.Molecules. 2022 May 3;27(9):2918. doi: 10.3390/molecules27092918. Molecules. 2022. PMID: 35566268 Free PMC article. Review.
-
Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation.PeerJ. 2021 Apr 13;9:e11261. doi: 10.7717/peerj.11261. eCollection 2021. PeerJ. 2021. PMID: 33954055 Free PMC article.
References
-
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
