Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target
- PMID: 32571877
- PMCID: PMC7450103
- DOI: 10.1074/jbc.RA120.013695
Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target
Abstract
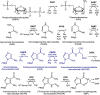
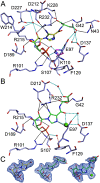
The bifunctional human enzyme phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase (PAICS) catalyzes two essential steps in the de novo purine biosynthesis pathway. PAICS is overexpressed in many cancers and could be a promising target for the development of cancer therapeutics. Here, using gene knockdowns and clonogenic survival and cell viability assays, we demonstrate that PAICS is required for growth and survival of prostate cancer cells. PAICS catalyzes the carboxylation of aminoimidazole ribonucleotide (AIR) and the subsequent conversion of carboxyaminoimidazole ribonucleotide (CAIR) and l-aspartate to N-succinylcarboxamide-5-aminoimidazole ribonucleotide (SAICAR). Of note, we present the first structures of human octameric PAICS in complexes with native ligands. In particular, we report the structure of PAICS with CAIR bound in the active sites of both domains and SAICAR bound in one of the SAICAR synthetase domains. Moreover, we report the PAICS structure with SAICAR and an ATP analog occupying the SAICAR synthetase active site. These structures provide insight into substrate and product binding and the architecture of the active sites, disclosing important structural information for rational design of PAICS inhibitors as potential anticancer drugs.
Keywords: N-succinylcarboxamide-5-aminoimidazole ribonucleotide (SAICAR); cancer target; cancer therapy; carboxyaminoimidazole ribonucleotide (CAIR); de novo purine biosynthesis; drug design; nucleoside/nucleotide biosynthesis; nucleotide metabolism; phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthetase (PAICS); prostate cancer; purine; rational drug design; structural biology.
© 2020 Škerlová et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures





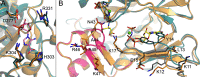
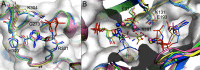
References
-
- Hartman S. C., and Buchanan J. M. (1959) Biosynthesis of the purines: XXVI. the identification of the formyl donors of the transformylation reactions. J. Biol. Chem. 234, 1812–1816 - PubMed
-
- Lukens L. N., and Buchanan J. M. (1959) Biosynthesis of the purines: XXIV. the enzymatic synthesis of 5-amino-1-ribosyl-4-imidazolecarboxylic acid 5'-phosphate from 5-amino-1-ribosylimidazole 5'-phosphate and carbon dioxide. J. Biol. Chem. 234, 1799–1805 - PubMed
-
- Barfeld S. J., Fazli L., Persson M., Marjavaara L., Urbanucci A., Kaukoniemi K. M., Rennie P. S., Ceder Y., Chabes A., Visakorpi T., and Mills I. G. (2015) Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget 6, 12587–12602 10.18632/oncotarget.3494 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

