Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium
- PMID: 32571970
- PMCID: PMC7948175
- DOI: 10.1136/gutjnl-2019-319919
Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium
Abstract
Objective: The epithelial layer of the GI tract is equipped with innate immune receptors to sense invading pathogens. Dysregulation in innate immune signalling pathways is associated with severe inflammatory diseases, but the responsiveness of GI epithelial cells to bacterial stimulation remains unclear.
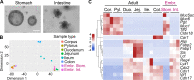
Design: We generated 42 lines of human and murine organoids from gastric and intestinal segments of both adult and fetal tissues. Genome-wide RNA-seq of the organoids provides an expression atlas of the GI epithelium. The innate immune response in epithelial cells was assessed using several functional assays in organoids and two-dimensional monolayers of cells from organoids.
Results: Results demonstrate extensive spatial organisation of innate immune signalling components along the cephalocaudal axis. A large part of this organisation is determined before birth and independent of exposure to commensal gut microbiota. Spatially restricted expression of Toll-like receptor 4 (Tlr4) in stomach and colon, but not in small intestine, is matched by nuclear factor kappa B (NF-κB) responses to lipopolysaccharide (LPS) exposure. Gastric epithelial organoids can sense LPS from the basal as well as from the apical side.
Conclusion: We conclude that the epithelial innate immune barrier follows a specific pattern per GI segment. The majority of the expression patterns and the function of TLR4 is encoded in the tissue-resident stem cells and determined primarily during development.
Keywords: epithelial barrier; gastrointestinal immune response; mucosal immunity.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
