Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States
- PMID: 32571980
- PMCID: PMC7319260
- DOI: 10.1126/scitranslmed.abc1126
Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States
Abstract
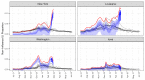
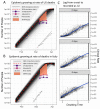
Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections to date has relied heavily on reverse transcription polymerase chain reaction testing. However, limited test availability, high false-negative rates, and the existence of asymptomatic or subclinical infections have resulted in an undercounting of the true prevalence of SARS-CoV-2. Here, we show how influenza-like illness (ILI) outpatient surveillance data can be used to estimate the prevalence of SARS-CoV-2. We found a surge of non-influenza ILI above the seasonal average in March 2020 and showed that this surge correlated with coronavirus disease 2019 (COVID-19) case counts across states. If one-third of patients infected with SARS-CoV-2 in the United States sought care, this ILI surge would have corresponded to more than 8.7 million new SARS-CoV-2 infections across the United States during the 3-week period from 8 to 28 March 2020. Combining excess ILI counts with the date of onset of community transmission in the United States, we also show that the early epidemic in the United States was unlikely to have been doubling slower than every 4 days. Together, these results suggest a conceptual model for the COVID-19 epidemic in the United States characterized by rapid spread across the United States with more than 80% infected individuals remaining undetected. We emphasize the importance of testing these findings with seroprevalence data and discuss the broader potential to use syndromic surveillance for early detection and understanding of emerging infectious diseases.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- World Health Organization, “Coronavirus disease 2019 (COVID-19): situation report, 59,” 2020.
-
- J. Lourenco, R. Paton, M. Ghafari, M. Kraemer, C. Thompson, P. Simmonds and S. Klenerman, “Fundamental principles of epidemic spread highlight the immediate need for large-scale serological surveys to assess the stage of the SARS-CoV-2 epidemic,” medRxiv, 2020.
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 395, 507–513 (2020). 10.1016/S0140-6736(20)30211-7 - DOI - PMC - PubMed
-
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z., Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020). 10.1001/jama.2020.1585 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

