Functional genomics identifies new synergistic therapies for retinoblastoma
- PMID: 32572160
- PMCID: PMC7391301
- DOI: 10.1038/s41388-020-1372-7
Functional genomics identifies new synergistic therapies for retinoblastoma
Abstract
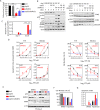
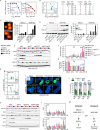
Local intravitreal or intra-arterial chemotherapy has improved therapeutic success for the pediatric cancer retinoblastoma (RB), but toxicity remains a major caveat. RB initiates primarily with RB1 loss or, rarely, MYCN amplification, but the critical downstream networks are incompletely understood. We set out to uncover perturbed molecular hubs, identify synergistic drug combinations to target these vulnerabilities, and expose and overcome drug resistance. We applied dynamic transcriptomic analysis to identify network hubs perturbed in RB versus normal fetal retina, and performed in vivo RNAi screens in RB1null and RB1wt;MYCNamp orthotopic xenografts to pinpoint essential hubs. We employed in vitro and in vivo studies to validate hits, define mechanism, develop new therapeutic modalities, and understand drug resistance. We identified BRCA1 and RAD51 as essential for RB cell survival. Their oncogenic activity was independent of BRCA1 functions in centrosome, heterochromatin, or ROS regulation, and instead linked to DNA repair. RAD51 depletion or inhibition with the small molecule inhibitor, B02, killed RB cells in a Chk1/Chk2/p53-dependent manner. B02 further synergized with clinically relevant topotecan (TPT) to engage this pathway, activating p53-BAX mediated killing of RB but not human retinal progenitor cells. Paradoxically, a B02/TPT-resistant tumor exhibited more DNA damage than sensitive RB cells. Resistance reflected dominance of the p53-p21 axis, which mediated cell cycle arrest instead of death. Deleting p21 or applying the BCL2/BCL2L1 inhibitor Navitoclax re-engaged the p53-BAX axis, and synergized with B02, TPT or both to override resistance. These data expose new synergistic therapies to trigger p53-induced killing in diverse RB subtypes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosom Cancer. 2007;46:617–34. - PubMed
-
- Rushlow DE, Mol BM, Kennett JY, Yee S, Pajovic S, Thériault BL, et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol. 2013;14:327–34. - PubMed
-
- Munier FL, Gaillard M-C, Balmer A, Soliman S, Podilsky G, Moulin AP, et al. Intravitreal chemotherapy for vitreous disease in retinoblastoma revisited: from prohibition to conditional indications. Br J Ophthalmol. 2012;96:1078–83. - PubMed
-
- Francis JH, Abramson DH, Gaillard M-C, Marr BP, Beck-Popovic M, Munier FL. The classification of vitreous seeds in retinoblastoma and response to intravitreal melphalan. Ophthalmology. 2015;122:1173–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

