Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity
- PMID: 32572240
- PMCID: PMC9291269
- DOI: 10.1038/s41590-020-0698-1
Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity
Abstract
The molecular basis for the propensity of a small number of environmental proteins to provoke allergic responses is largely unknown. Herein, we report that mite group 13 allergens of the fatty acid-binding protein (FABP) family are sensed by an evolutionarily conserved acute-phase protein, serum amyloid A1 (SAA1), that promotes pulmonary type 2 immunity. Mechanistically, SAA1 interacted directly with allergenic mite FABPs (Der p 13 and Blo t 13). The interaction between mite FABPs and SAA1 activated the SAA1-binding receptor, formyl peptide receptor 2 (FPR2), which drove the epithelial release of the type-2-promoting cytokine interleukin (IL)-33 in a SAA1-dependent manner. Importantly, the SAA1-FPR2-IL-33 axis was upregulated in nasal epithelial cells from patients with chronic rhinosinusitis. These findings identify an unrecognized role for SAA1 as a soluble pattern recognition receptor for conserved FABPs found in common mite allergens that initiate type 2 immunity at mucosal surfaces.
Conflict of interest statement
Figures


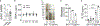
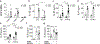

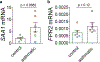
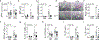
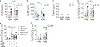
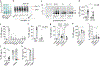
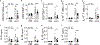


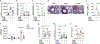
Comment in
-
A soluble allergen sensor sounds the alarm.Nat Immunol. 2020 Jul;21(7):724-726. doi: 10.1038/s41590-020-0709-2. Nat Immunol. 2020. PMID: 32572239 No abstract available.
References
-
- Corrigan CJ et al. Allergen-induced expression of IL-25 and IL-25 receptor in atopic asthmatic airways and late-phase cutaneous responses. J. Allergy Clin. Immunol 128, 116–124 (2011). - PubMed
-
- Schuijs MJ et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015). - PubMed
Methods only references
-
- Darveau ME, Jacques E, Rouabhia M, Hamid Q & Chakir J Increased T-cell survival by structural bronchial cells derived from asthmatic subjects cultured in an engineered human mucosa. J. Allergy Clin. Immunol 121, 692–699 (2008). - PubMed
-
- Goulet F et al. Morphologic and functional properties of bronchial cells isolated from normal and asthmatic subjects. Am. J. Respir. Cell Mol. Biol 15, 312–318 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

