Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes
- PMID: 32572265
- PMCID: PMC8500275
- DOI: 10.1038/s41591-020-0932-2
Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes
Abstract
Definitive diagnosis of intracranial tumors relies on tissue specimens obtained by invasive surgery. Noninvasive diagnostic approaches provide an opportunity to avoid surgery and mitigate unnecessary risk to patients. In the present study, we show that DNA-methylation profiles from plasma reveal highly specific signatures to detect and accurately discriminate common primary intracranial tumors that share cell-of-origin lineages and can be challenging to distinguish using standard-of-care imaging.
Conflict of interest statement
Competing interests
D.D.D.C., S.Y.S. and A.C. are listed as inventors on patents filed that are related to this method. D.D.D.C. received research funding from Pfizer and Nektar therapeutics not related to this project.
Figures
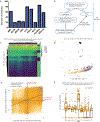
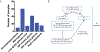


Comment in
-
Blood Test Catches Cancers That Shed Little DNA.Cancer Discov. 2020 Sep;10(9):1246-1247. doi: 10.1158/2159-8290.CD-NB2020-068. Epub 2020 Jul 14. Cancer Discov. 2020. PMID: 32665301
-
Closing in on cfDNA-based detection and diagnosis.Nat Rev Cancer. 2020 Sep;20(9):481. doi: 10.1038/s41568-020-0293-7. Nat Rev Cancer. 2020. PMID: 32690930 No abstract available.
-
Methylation extends the reach of liquid biopsy in cancer detection.Nat Rev Clin Oncol. 2020 Nov;17(11):655-656. doi: 10.1038/s41571-020-0420-0. Nat Rev Clin Oncol. 2020. PMID: 32732909 Free PMC article.
References
-
- Heitzer E, Haque IS, Roberts CES & Speicher MR Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019). - PubMed
-
- Shen SY et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). - PubMed
-
- Shen SY, Burgener JM, Bratman SV & De Carvalho DD Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc. 14, 2749–2780 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

