A role of the frontotemporal lobar degeneration risk factor TMEM106B in myelination
- PMID: 32572497
- PMCID: PMC7363491
- DOI: 10.1093/brain/awaa154
A role of the frontotemporal lobar degeneration risk factor TMEM106B in myelination
Abstract
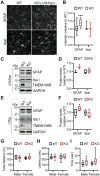
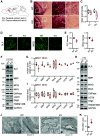
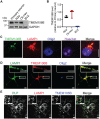
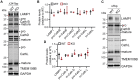
TMEM106B encodes a lysosomal membrane protein and was initially identified as a risk factor for frontotemporal lobar degeneration. Recently, a dominant D252N mutation in TMEM106B was shown to cause hypomyelinating leukodystrophy. However, how TMEM106B regulates myelination is still unclear. Here we show that TMEM106B is expressed and localized to the lysosome compartment in oligodendrocytes. TMEM106B deficiency in mice results in myelination defects with a significant reduction of protein levels of proteolipid protein (PLP) and myelin oligodendrocyte glycoprotein (MOG), the membrane proteins found in the myelin sheath. The levels of many lysosome proteins are significantly decreased in the TMEM106B-deficient Oli-neu oligodendroglial precursor cell line. TMEM106B physically interacts with the lysosomal protease cathepsin D and is required to maintain proper cathepsin D levels in oligodendrocytes. Furthermore, we found that TMEM106B deficiency results in lysosome clustering in the perinuclear region and a decrease in lysosome exocytosis and cell surface PLP levels. Moreover, we found that the D252N mutation abolished lysosome enlargement and lysosome acidification induced by wild-type TMEM106B overexpression. Instead, it stimulates lysosome clustering near the nucleus as seen in TMEM106B-deficient cells. Our results support that TMEM106B regulates myelination through modulation of lysosome function in oligodendrocytes.
Keywords: TMEM106B; lysosome; myelination; oligodendrocytes.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Superresolution live-cell imaging reveals that the localization of TMEM106B to filopodia in oligodendrocytes is compromised by the hypomyelination-related D252N mutation.Sci China Life Sci. 2023 Aug;66(8):1858-1868. doi: 10.1007/s11427-022-2290-1. Epub 2023 Apr 18. Sci China Life Sci. 2023. PMID: 37129766
-
Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency.Acta Neuropathol Commun. 2017 Jan 26;5(1):9. doi: 10.1186/s40478-017-0412-1. Acta Neuropathol Commun. 2017. PMID: 28126008 Free PMC article.
-
The FTLD Risk Factor TMEM106B Regulates the Transport of Lysosomes at the Axon Initial Segment of Motoneurons.Cell Rep. 2020 Mar 10;30(10):3506-3519.e6. doi: 10.1016/j.celrep.2020.02.060. Cell Rep. 2020. PMID: 32160553
-
Physiological and pathological functions of TMEM106B: a gene associated with brain aging and multiple brain disorders.Acta Neuropathol. 2021 Mar;141(3):327-339. doi: 10.1007/s00401-020-02246-3. Epub 2021 Jan 1. Acta Neuropathol. 2021. PMID: 33386471 Free PMC article. Review.
-
Identification of TMEM106B amyloid fibrils provides an updated view of TMEM106B biology in health and disease.Acta Neuropathol. 2022 Nov;144(5):807-819. doi: 10.1007/s00401-022-02486-5. Epub 2022 Sep 2. Acta Neuropathol. 2022. PMID: 36056242 Free PMC article. Review.
Cited by
-
Emerging cellular themes in leukodystrophies.Front Cell Dev Biol. 2022 Aug 8;10:902261. doi: 10.3389/fcell.2022.902261. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36003149 Free PMC article. Review.
-
The TMEM106B T186S coding variant increases neurite arborization and synaptic density in primary hippocampal neurons.Front Neurosci. 2023 Oct 13;17:1275959. doi: 10.3389/fnins.2023.1275959. eCollection 2023. Front Neurosci. 2023. PMID: 37901434 Free PMC article.
-
Myristoylation of TMEM106B by NMT1/2 regulates TMEM106B trafficking and turnover.J Biol Chem. 2025 Jul;301(7):110322. doi: 10.1016/j.jbc.2025.110322. Epub 2025 May 30. J Biol Chem. 2025. PMID: 40451428 Free PMC article.
-
TMEM106B aggregation in neurodegenerative diseases: linking genetics to function.Mol Neurodegener. 2023 Aug 10;18(1):54. doi: 10.1186/s13024-023-00644-1. Mol Neurodegener. 2023. PMID: 37563705 Free PMC article. Review.
-
White matter pathology in FTLD caused by GRN mutations.Acta Neuropathol. 2025 Jan 15;149(1):7. doi: 10.1007/s00401-025-02847-w. Acta Neuropathol. 2025. PMID: 39812821 No abstract available.
References
-
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F.. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol 1990; 27: 229–37. - PubMed
-
- Charzewska A, Wierzba J, Izycka-Swieszewska E, Bekiesinska-Figatowska M, Jurek M, Gintowt A, et al.Hypomyelinating leukodystrophies - a molecular insight into the white matter pathology. Clin Genet 2016; 90: 293–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

