Three Copies of Four Interferon Receptor Genes Underlie a Mild Type I Interferonopathy in Down Syndrome
- PMID: 32572726
- PMCID: PMC7418179
- DOI: 10.1007/s10875-020-00803-9
Three Copies of Four Interferon Receptor Genes Underlie a Mild Type I Interferonopathy in Down Syndrome
Abstract
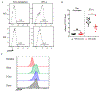
Down syndrome (DS) is characterized by the occurrence of three copies of human chromosome 21 (HSA21). HSA21 contains a cluster of four interferon receptor (IFN-R) genes: IFNAR1, IFNAR2, IFNGR2, and IL10RB. DS patients often develop mucocutaneous infections and autoimmune diseases, mimicking patients with heterozygous gain-of-function (GOF) STAT1 mutations, which enhance cellular responses to three types of interferon (IFN). A gene dosage effect at these four loci may contribute to the infectious and autoimmune manifestations observed in individuals with DS. We report high levels of IFN-αR1, IFN-αR2, and IFN-γR2 expression on the surface of monocytes and EBV-transformed-B (EBV-B) cells from studying 45 DS patients. Total and phosphorylated STAT1 (STAT1 and pSTAT1) levels were constitutively high in unstimulated and IFN-α- and IFN-γ-stimulated monocytes from DS patients but lower than those in patients with GOF STAT1 mutations. Following stimulation with IFN-α or -γ, but not with IL-6 or IL-21, pSTAT1 and IFN-γ activation factor (GAF) DNA-binding activities were significantly higher in the EBV-B cells of DS patients than in controls. These responses resemble the dysregulated responses observed in patients with STAT1 GOF mutations. Concentrations of plasma type I IFNs were high in 12% of the DS patients tested (1.8% in the healthy controls). Levels of type I IFNs, IFN-Rs, and STAT1 were similar in DS patients with and without recurrent skin infections. We performed a genome-wide transcriptomic analysis based on principal component analysis and interferon modules on circulating monocytes. We found that DS monocytes had levels of both IFN-α- and IFN-γ-inducible ISGs intermediate to those of monocytes from healthy controls and from patients with GOF STAT1 mutations. Unlike patients with GOF STAT1 mutations, patients with DS had normal circulating Th17 counts and a high proportion of terminally differentiated CD8+ T cells with low levels of STAT1 expression. We conclude a mild interferonopathy in Down syndrome leads to an incomplete penetrance at both cellular and clinical level, which is not correlate with recurrent skin bacterial or fungal infections. The constitutive upregulation of type I and type II IFN-R, at least in monocytes of DS patients, may contribute to the autoimmune diseases observed in these individuals.
Keywords: Down syndrome; JAK-STAT; interferon receptors; interferonopathy.
Conflict of interest statement
Conflict of Interest:
Dr. Casanova reports personal fees from Celgene, personal fees from ADMA, personal fees from Nimbus, personal fees from Vitae Pharmaceuticals, Inc, personal fees from KymeraTX, personal fees from Sanofi, personal fees from Asahi Kasei, personal fees from Pfizer, personal fees from Elixiron Immunotherapeutics, outside the submitted work. None of the other authors have no conflict of interest to declare.
Figures


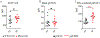
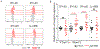


References
-
- Lejeune J, Gautier M & Turpin R A study of somatic chromosomes in nine infants with mongolism. C.R. Acad. Sci 248, 1721–1722 (1959). - PubMed
-
- de Graaf G, Buckley F & Skotko BG Estimation of the number of people with Down syndrome in the United States. Genet. Med 19, 439–447 (2017). - PubMed
-
- Oster J, Mikkelsen M & Nielsen A Mortality and life-table in Down’s syndrome. Acta Paediatr. Scand 64, 322–6 (1975). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

