Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach
- PMID: 32573351
- PMCID: PMC7332876
- DOI: 10.1080/07391102.2020.1778537
Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach
Abstract
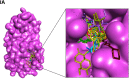
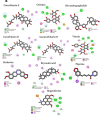
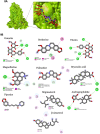
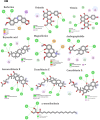
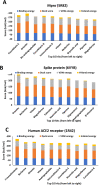
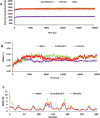
In the present study, we have explored the interaction of the active components from 10 different medicinal plants of Indian origin that are commonly used for treating cold and respiratory-related disorders, through molecular docking analysis. In the current scenario, COVID-19 patients experience severe respiratory syndromes, hence it is envisaged from our study that these traditional medicines are very likely to provide a favourable effect on COVID-19 infections. The active ingredients identified from these natural products are previously reported for antiviral activities against large group of viruses. Totally 47 bioactives identified from the medicinal plants were investigated against the structural targets of SARS-CoV-2 (Mpro and spike protein) and human ACE2 receptor. The top leads were identified based on interaction energies, number of hydrogen bond and other parameters that explain their potency to inhibit SARS-CoV-2. The bioactive ligands such as Cucurbitacin E, Orientin, Bis-andrographolide, Cucurbitacin B, Isocucurbitacin B, Vitexin, Berberine, Bryonolic acid, Piperine and Magnoflorine targeted the hotspot residues of SARS-CoV-2 main protease. In fact, this protease enzyme has an essential role in mediating the viral replication and therefore compounds targeting this key enzyme are expected to block the viral replication and transcription. The top scoring conformations identified through docking analysis were further demonstrated with molecular dynamics simulation. Besides, the stability of the conformation was studied in detail by investigating the binding free energy using MM-PBSA method. Overall, the study emphasized that the proposed hit Cucurbitacin E and orientin could serve as a promising scaffold for developing anti-COVID-19 drug.Communicated by Ramaswamy H. Sarma.
Keywords: Binding energy; Ethnomedicine; MD simulation; SARS-CoV-2 inhibitors; drug discovery; molecular docking.
Figures


References
-
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 - DOI - PMC - PubMed
-
- Adeoye A. O., Oso B. J., Olaoye I. F., Tijjani H., & Adebayo A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1765876. - DOI - PMC - PubMed
-
- Alsayari A., Darweesh M., Halaweish F., & Chase C. C. L. (2012). Anti-bovine viral diarrhea virus activity of cucurbitacins as new potential antiviral agents. Planta Medica, 78(11), PD142 10.1055/s-0032-1320500 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
