BioModels Parameters: a treasure trove of parameter values from published systems biology models
- PMID: 32573648
- PMCID: PMC7653554
- DOI: 10.1093/bioinformatics/btaa560
BioModels Parameters: a treasure trove of parameter values from published systems biology models
Abstract
Motivation: One of the major bottlenecks in building systems biology models is identification and estimation of model parameters for model calibration. Searching for model parameters from published literature and models is an essential, yet laborious task.
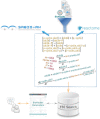
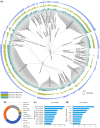
Results: We have developed a new service, BioModels Parameters, to facilitate search and retrieval of parameter values from the Systems Biology Markup Language models stored in BioModels. Modellers can now directly search for a model entity (e.g. a protein or drug) to retrieve the rate equations describing it; the associated parameter values (e.g. degradation rate, production rate, Kcat, Michaelis-Menten constant, etc.) and the initial concentrations. Currently, BioModels Parameters contains entries from over 84,000 reactions and 60 different taxa with cross-references. The retrieved rate equations and parameters can be used for scanning parameter ranges, model fitting and model extension. Thus, BioModels Parameters will be a valuable service for systems biology modellers.
Availability and implementation: The data are accessible via web interface and API. BioModels Parameters is free to use and is publicly available at https://www.ebi.ac.uk/biomodels/parameterSearch.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures


References
-
- Aldridge B.B. et al. (2006) Physicochemical modelling of cell signalling pathways. Nat. Cell Biol., 8, 1195–1203. - PubMed
-
- Bungay S.D. et al. (2006) Modelling thrombin generation in human ovarian follicular fluid. Bull. Math. Biol., 68, 2283–2302. - PubMed
-
- Hoops S. et al. (2006) COPASI—a COmplex PAthway SImulator. Bioinformatics, 22, 3067–3074. - PubMed

