Distinct subtypes of polycystic ovary syndrome with novel genetic associations: An unsupervised, phenotypic clustering analysis
- PMID: 32574161
- PMCID: PMC7310679
- DOI: 10.1371/journal.pmed.1003132
Distinct subtypes of polycystic ovary syndrome with novel genetic associations: An unsupervised, phenotypic clustering analysis
Abstract
Background: Polycystic ovary syndrome (PCOS) is a common, complex genetic disorder affecting up to 15% of reproductive-age women worldwide, depending on the diagnostic criteria applied. These diagnostic criteria are based on expert opinion and have been the subject of considerable controversy. The phenotypic variation observed in PCOS is suggestive of an underlying genetic heterogeneity, but a recent meta-analysis of European ancestry PCOS cases found that the genetic architecture of PCOS defined by different diagnostic criteria was generally similar, suggesting that the criteria do not identify biologically distinct disease subtypes. We performed this study to test the hypothesis that there are biologically relevant subtypes of PCOS.
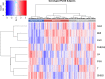
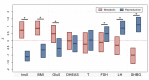
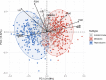
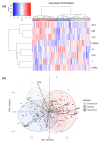
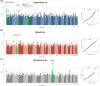
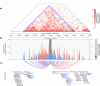
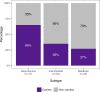
Methods and findings: Using biochemical and genotype data from a previously published PCOS genome-wide association study (GWAS), we investigated whether there were reproducible phenotypic subtypes of PCOS with subtype-specific genetic associations. Unsupervised hierarchical cluster analysis was performed on quantitative anthropometric, reproductive, and metabolic traits in a genotyped cohort of 893 PCOS cases (median and interquartile range [IQR]: age = 28 [25-32], body mass index [BMI] = 35.4 [28.2-41.5]). The clusters were replicated in an independent, ungenotyped cohort of 263 PCOS cases (median and IQR: age = 28 [24-33], BMI = 35.7 [28.4-42.3]). The clustering revealed 2 distinct PCOS subtypes: a "reproductive" group (21%-23%), characterized by higher luteinizing hormone (LH) and sex hormone binding globulin (SHBG) levels with relatively low BMI and insulin levels, and a "metabolic" group (37%-39%), characterized by higher BMI, glucose, and insulin levels with lower SHBG and LH levels. We performed a GWAS on the genotyped cohort, limiting the cases to either the reproductive or metabolic subtypes. We identified alleles in 4 loci that were associated with the reproductive subtype at genome-wide significance (PRDM2/KAZN, P = 2.2 × 10-10; IQCA1, P = 2.8 × 10-9; BMPR1B/UNC5C, P = 9.7 × 10-9; CDH10, P = 1.2 × 10-8) and one locus that was significantly associated with the metabolic subtype (KCNH7/FIGN, P = 1.0 × 10-8). We developed a predictive model to classify a separate, family-based cohort of 73 women with PCOS (median and IQR: age = 28 [25-33], BMI = 34.3 [27.8-42.3]) and found that the subtypes tended to cluster in families and that carriers of previously reported rare variants in DENND1A, a gene that regulates androgen biosynthesis, were significantly more likely to have the reproductive subtype of PCOS. Limitations of our study were that only PCOS cases of European ancestry diagnosed by National Institutes of Health (NIH) criteria were included, the sample sizes for the subtype GWAS were small, and the GWAS findings were not replicated.
Conclusions: In conclusion, we have found reproducible reproductive and metabolic subtypes of PCOS. Furthermore, these subtypes were associated with novel, to our knowledge, susceptibility loci. Our results suggest that these subtypes are biologically relevant because they appear to have distinct genetic architecture. This study demonstrates how phenotypic subtyping can be used to gain additional insights from GWAS data.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: GNN owns equity in RenalytixAI, Inc., and receives financial compensation as a consultant and advisory board member for RenalytixAI. GNN has received operational funding from Goldfinch Bio and consulting fees from BioVie Inc. and GLG consulting in the past three years. GNN is a former member of the advisory board of PulseData and received consulting fees for their services and continues to hold equity interests in PulseData.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

