Development of small-molecule inhibitors of fatty acyl-AMP and fatty acyl-CoA ligases in Mycobacterium tuberculosis
- PMID: 32574901
- PMCID: PMC7415619
- DOI: 10.1016/j.ejmech.2020.112408
Development of small-molecule inhibitors of fatty acyl-AMP and fatty acyl-CoA ligases in Mycobacterium tuberculosis
Abstract
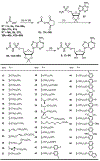
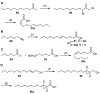
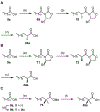
Lipid metabolism in Mycobacterium tuberculosis (Mtb) relies on 34 fatty acid adenylating enzymes (FadDs) that can be grouped into two classes: fatty acyl-CoA ligases (FACLs) involved in lipid and cholesterol catabolism and long chain fatty acyl-AMP ligases (FAALs) involved in biosynthesis of the numerous essential and virulence-conferring lipids found in Mtb. The precise biochemical roles of many FACLs remain poorly characterized while the functionally non-redundant FAALs are much better understood. Here we describe the systematic investigation of 5'-O-[N-(alkanoyl)sulfamoyl]adenosine (alkanoyl adenosine monosulfamate, alkanoyl-AMS) analogs as potential multitarget FadD inhibitors for their antitubercular activity and biochemical selectivity towards representative FAAL and FACL enzymes. We identified several potent compounds including 12-azidododecanoyl-AMS 28, 11-phenoxyundecanoyl-AMS 32, and nonyloxyacetyl-AMS 36 with minimum inhibitory concentrations (MICs) against M. tuberculosis ranging from 0.098 to 3.13 μM. Compound 32 was notable for its impressive biochemical selectivity against FAAL28 (apparent Ki = 0.7 μM) versus FACL19 (Ki > 100 μM), and uniform activity against a panel of multidrug and extensively drug-resistant TB strains with MICs ranging from 3.13 to 12.5 μM in minimal (GAST) and rich (7H9) media. The SAR analysis provided valuable insights for further optimization of 32 and also identified limitations to overcome.
Keywords: Acyl-AMS analogs; FAAL28; FACL19; Fatty acyl-AMP ligases; Fatty acyl-CoA ligases; Mycobacterium tuberculosis.
Copyright © 2020 Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

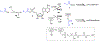
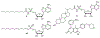
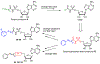

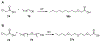

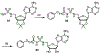

References
-
- WHO 2019 Global Tuberculosis Report, World Health Organization: France, 2019; Chapter 3, pp 27–70. https://apps.who.int/iris/handle/10665/329368 (accessed April 4, 2020).
-
- WHO 2019 consolidated guidelines on drug-resistant tuberculosis treatment, World Health Organization: Switzerland, 2019; pp 19–41. https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-re... (accessed April 4, 2020). - PubMed
-
- Gokhale RS, Saxena P, Chopra T, Mohanty D, Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids, Nat Prod Rep, 24 (2007) 267–277. - PubMed
-
- Quadri LE, Biosynthesis of mycobacterial lipids by polyketide synthases and beyond, Critical reviews in biochemistry and molecular biology, 49 (2014) 179–211. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Miscellaneous

